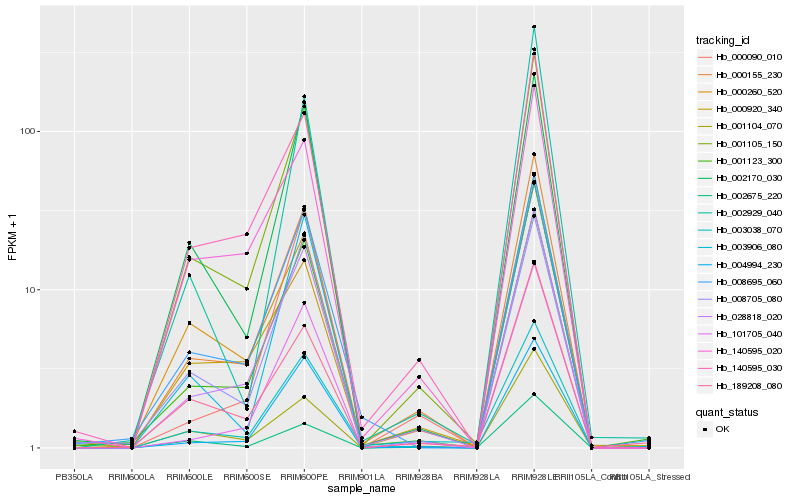
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000155_230 |
0.0 |
- |
- |
rac gtpase, putative [Ricinus communis] |
| 2 |
Hb_001105_150 |
0.0397570978 |
- |
- |
GERMIN-LIKE protein 10 [Populus trichocarpa] |
| 3 |
Hb_028818_020 |
0.0536764586 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9-like [Jatropha curcas] |
| 4 |
Hb_140595_030 |
0.0659023931 |
- |
- |
- |
| 5 |
Hb_001123_300 |
0.0809780219 |
- |
- |
hypothetical protein POPTR_0003s16270g [Populus trichocarpa] |
| 6 |
Hb_101705_040 |
0.0840786855 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 7 |
Hb_140595_020 |
0.0863132577 |
- |
- |
PREDICTED: vegetative cell wall protein gp1-like [Solanum lycopersicum] |
| 8 |
Hb_189208_080 |
0.0944903971 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter B family member 11-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_008695_060 |
0.0957097256 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 10 |
Hb_003038_070 |
0.0965020067 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 11 |
Hb_001104_070 |
0.0974713553 |
- |
- |
hypothetical protein SORBIDRAFT_10g024640 [Sorghum bicolor] |
| 12 |
Hb_000260_520 |
0.1003580785 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 13 |
Hb_002675_220 |
0.1020992012 |
- |
- |
PREDICTED: probable pyridoxal biosynthesis protein PDX1.2 [Populus euphratica] |
| 14 |
Hb_000920_340 |
0.1050329663 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 15 |
Hb_004994_230 |
0.1055235369 |
transcription factor |
TF Family: LIM |
Pollen-specific protein SF3, putative [Ricinus communis] |
| 16 |
Hb_002170_030 |
0.1081241334 |
- |
- |
PREDICTED: laccase-4-like [Jatropha curcas] |
| 17 |
Hb_000090_010 |
0.1110199027 |
- |
- |
hypothetical protein JCGZ_22567 [Jatropha curcas] |
| 18 |
Hb_003906_080 |
0.1115707241 |
- |
- |
Peroxidase 66 precursor, putative [Ricinus communis] |
| 19 |
Hb_008705_080 |
0.1138389623 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 14 [Jatropha curcas] |
| 20 |
Hb_002929_040 |
0.1142134708 |
- |
- |
zinc finger protein, putative [Ricinus communis] |