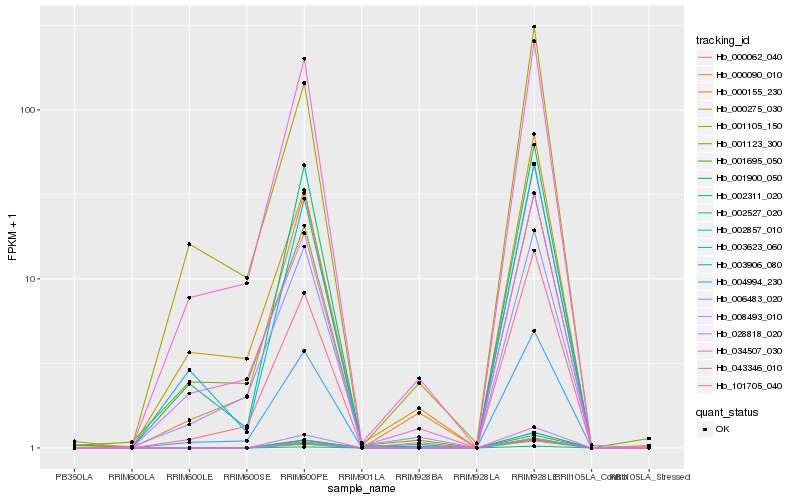
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_101705_040 |
0.0 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 2 |
Hb_000090_010 |
0.0555741439 |
- |
- |
hypothetical protein JCGZ_22567 [Jatropha curcas] |
| 3 |
Hb_004994_230 |
0.0636675404 |
transcription factor |
TF Family: LIM |
Pollen-specific protein SF3, putative [Ricinus communis] |
| 4 |
Hb_028818_020 |
0.0761844914 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9-like [Jatropha curcas] |
| 5 |
Hb_000155_230 |
0.0840786855 |
- |
- |
rac gtpase, putative [Ricinus communis] |
| 6 |
Hb_034507_030 |
0.0903340215 |
- |
- |
PREDICTED: laccase-4-like [Jatropha curcas] |
| 7 |
Hb_001105_150 |
0.0906979551 |
- |
- |
GERMIN-LIKE protein 10 [Populus trichocarpa] |
| 8 |
Hb_003906_080 |
0.1007911511 |
- |
- |
Peroxidase 66 precursor, putative [Ricinus communis] |
| 9 |
Hb_006483_020 |
0.1012528091 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_001123_300 |
0.1030730215 |
- |
- |
hypothetical protein POPTR_0003s16270g [Populus trichocarpa] |
| 11 |
Hb_002311_020 |
0.1052685021 |
- |
- |
PREDICTED: putative beta-D-xylosidase [Jatropha curcas] |
| 12 |
Hb_003623_060 |
0.112568315 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000062_040 |
0.1125714306 |
- |
- |
PREDICTED: uncharacterized protein LOC105766209 [Gossypium raimondii] |
| 14 |
Hb_000275_030 |
0.1130173128 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001900_050 |
0.1133927619 |
- |
- |
PREDICTED: uncharacterized protein LOC102609547 [Citrus sinensis] |
| 16 |
Hb_002527_020 |
0.1135307747 |
- |
- |
PREDICTED: F-box protein At1g30790-like [Jatropha curcas] |
| 17 |
Hb_002857_010 |
0.113736703 |
- |
- |
leucine-rich repeat transmembrane protein kinase [Populus trichocarpa] |
| 18 |
Hb_043346_010 |
0.1140717174 |
- |
- |
PREDICTED: uncharacterized protein LOC105761922 [Gossypium raimondii] |
| 19 |
Hb_001695_050 |
0.1145777536 |
- |
- |
PREDICTED: uncharacterized protein LOC104884077 [Beta vulgaris subsp. vulgaris] |
| 20 |
Hb_008493_010 |
0.116094081 |
- |
- |
hypothetical protein JCGZ_10529 [Jatropha curcas] |