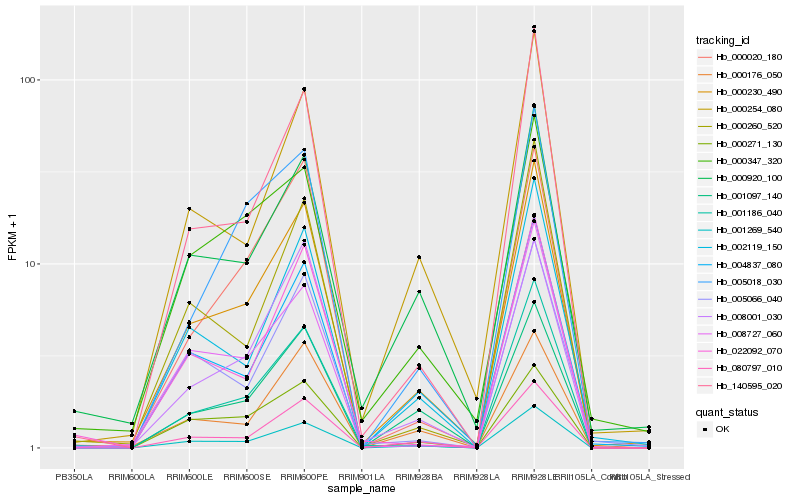
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000230_490 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001269_540 |
0.0616944103 |
- |
- |
hypothetical protein VITISV_038016 [Vitis vinifera] |
| 3 |
Hb_080797_010 |
0.0820658543 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570-like [Citrus sinensis] |
| 4 |
Hb_004837_080 |
0.0847037346 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB46 [Jatropha curcas] |
| 5 |
Hb_001186_040 |
0.0913912348 |
- |
- |
Peroxidase 3 precursor, putative [Ricinus communis] |
| 6 |
Hb_140595_020 |
0.092560425 |
- |
- |
PREDICTED: vegetative cell wall protein gp1-like [Solanum lycopersicum] |
| 7 |
Hb_002119_150 |
0.095251103 |
- |
- |
hypothetical protein POPTR_0008s13870g [Populus trichocarpa] |
| 8 |
Hb_000254_080 |
0.0959414558 |
- |
- |
PREDICTED: uncharacterized protein LOC105635000 [Jatropha curcas] |
| 9 |
Hb_000920_100 |
0.0980684776 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 10 |
Hb_005066_040 |
0.1094304165 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 11 |
Hb_001097_140 |
0.1094858985 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000260_520 |
0.1114593238 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 13 |
Hb_000176_050 |
0.1115475142 |
- |
- |
glutamine amidotransferase class-I domain-containing family protein [Populus trichocarpa] |
| 14 |
Hb_000347_320 |
0.1122992665 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 15 |
Hb_008001_030 |
0.114862211 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000020_180 |
0.1149437808 |
- |
- |
laccase, putative [Ricinus communis] |
| 17 |
Hb_008727_060 |
0.1151176323 |
- |
- |
Beta-glucosidase, putative [Ricinus communis] |
| 18 |
Hb_022092_070 |
0.1161585143 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.5 [Jatropha curcas] |
| 19 |
Hb_000271_130 |
0.1184769258 |
transcription factor |
TF Family: C2C2-Dof |
DNA binding with one finger 2.4, putative [Theobroma cacao] |
| 20 |
Hb_005018_030 |
0.1188934057 |
- |
- |
hypothetical protein CICLE_v10019811mg [Citrus clementina] |