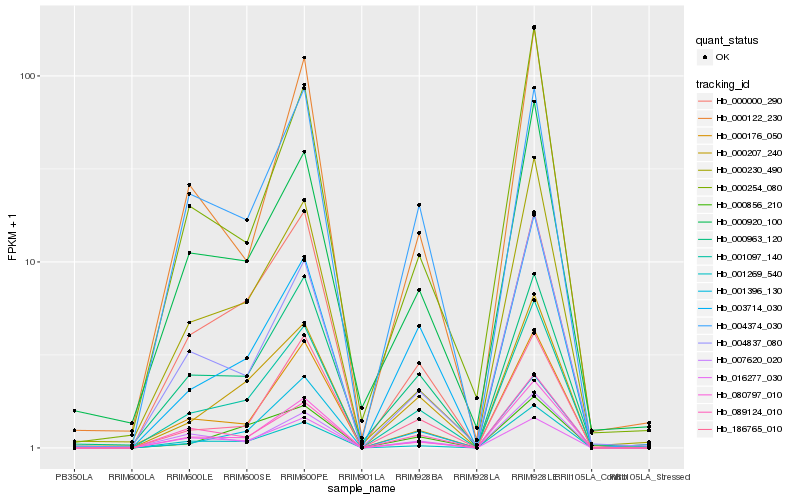
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001097_140 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_080797_010 |
0.0710364655 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570-like [Citrus sinensis] |
| 3 |
Hb_000207_240 |
0.0734690776 |
- |
- |
hypothetical protein RCOM_0686540 [Ricinus communis] |
| 4 |
Hb_003714_030 |
0.0966469321 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 5 |
Hb_001269_540 |
0.0986824471 |
- |
- |
hypothetical protein VITISV_038016 [Vitis vinifera] |
| 6 |
Hb_186765_010 |
0.0988640199 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105629453 [Jatropha curcas] |
| 7 |
Hb_004837_080 |
0.1041926198 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB46 [Jatropha curcas] |
| 8 |
Hb_089124_010 |
0.1046646708 |
- |
- |
hypothetical protein POPTR_0018s09540g [Populus trichocarpa] |
| 9 |
Hb_000176_050 |
0.1061971251 |
- |
- |
glutamine amidotransferase class-I domain-containing family protein [Populus trichocarpa] |
| 10 |
Hb_000000_290 |
0.1067942517 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 4 [Jatropha curcas] |
| 11 |
Hb_000920_100 |
0.1092979259 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 12 |
Hb_000963_120 |
0.1094776055 |
- |
- |
PREDICTED: uncharacterized protein LOC105628103 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000230_490 |
0.1094858985 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001396_130 |
0.1103003862 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_007620_020 |
0.1112259518 |
- |
- |
PREDICTED: cationic amino acid transporter 7, chloroplastic-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000122_230 |
0.1177497487 |
- |
- |
laccase, putative [Ricinus communis] |
| 17 |
Hb_004374_030 |
0.1211148472 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 18 |
Hb_016277_030 |
0.12446403 |
- |
- |
hypothetical protein JCGZ_16973 [Jatropha curcas] |
| 19 |
Hb_000254_080 |
0.1245428813 |
- |
- |
PREDICTED: uncharacterized protein LOC105635000 [Jatropha curcas] |
| 20 |
Hb_000856_210 |
0.1254339074 |
- |
- |
nitrate transporter, putative [Ricinus communis] |