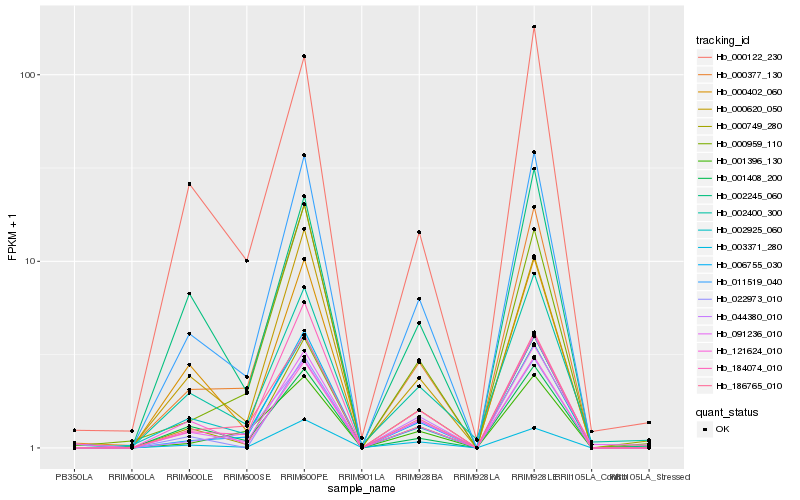
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011519_040 |
0.0 |
- |
- |
hypothetical protein POPTR_0015s04770g [Populus trichocarpa] |
| 2 |
Hb_000377_130 |
0.0642143349 |
- |
- |
hypothetical protein POPTR_0016s00790g [Populus trichocarpa] |
| 3 |
Hb_186765_010 |
0.0671345273 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105629453 [Jatropha curcas] |
| 4 |
Hb_006755_030 |
0.0728225101 |
- |
- |
BURP domain-containing protein, putative [Theobroma cacao] |
| 5 |
Hb_091236_010 |
0.0865348956 |
- |
- |
hypothetical protein POPTR_0019s12290g [Populus trichocarpa] |
| 6 |
Hb_000620_050 |
0.0930286044 |
- |
- |
PREDICTED: uncharacterized protein LOC105649924 [Jatropha curcas] |
| 7 |
Hb_001408_200 |
0.0980823705 |
- |
- |
hypothetical protein JCGZ_08964 [Jatropha curcas] |
| 8 |
Hb_002925_060 |
0.0982517181 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01547 [Jatropha curcas] |
| 9 |
Hb_000749_280 |
0.0992447988 |
- |
- |
PREDICTED: uncharacterized protein LOC105138169 [Populus euphratica] |
| 10 |
Hb_184074_010 |
0.1138760854 |
- |
- |
PREDICTED: uncharacterized protein LOC105639054 [Jatropha curcas] |
| 11 |
Hb_044380_010 |
0.1142769806 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Malus domestica] |
| 12 |
Hb_000122_230 |
0.1144465948 |
- |
- |
laccase, putative [Ricinus communis] |
| 13 |
Hb_121624_010 |
0.1147581696 |
- |
- |
PREDICTED: uncharacterized protein LOC105634410 [Jatropha curcas] |
| 14 |
Hb_000402_060 |
0.114776386 |
- |
- |
PREDICTED: uncharacterized protein LOC105644642 isoform X2 [Jatropha curcas] |
| 15 |
Hb_002245_060 |
0.1160211073 |
- |
- |
PREDICTED: kinesin-3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003371_280 |
0.1163030835 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_000959_110 |
0.1198487478 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 13 [Vitis vinifera] |
| 18 |
Hb_002400_300 |
0.12169056 |
- |
- |
PREDICTED: synaptotagmin-4 [Jatropha curcas] |
| 19 |
Hb_022973_010 |
0.1223288626 |
- |
- |
PREDICTED: trans-resveratrol di-O-methyltransferase-like [Jatropha curcas] |
| 20 |
Hb_001396_130 |
0.1224994461 |
- |
- |
conserved hypothetical protein [Ricinus communis] |