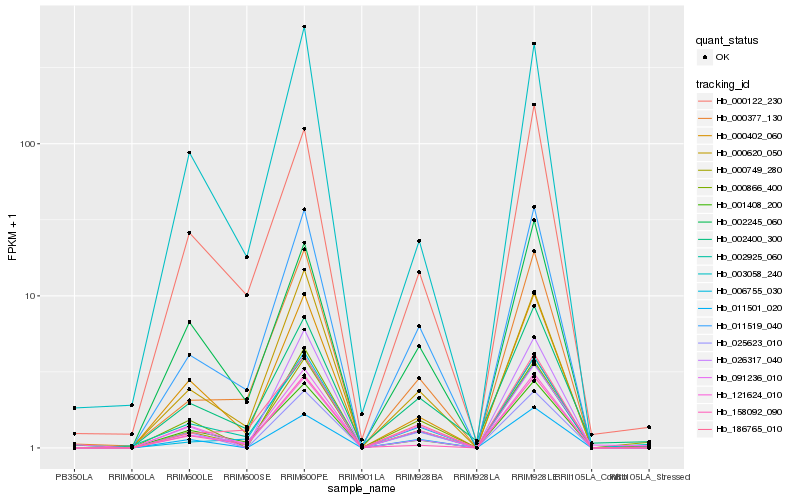
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000749_280 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105138169 [Populus euphratica] |
| 2 |
Hb_091236_010 |
0.0907589463 |
- |
- |
hypothetical protein POPTR_0019s12290g [Populus trichocarpa] |
| 3 |
Hb_011519_040 |
0.0992447988 |
- |
- |
hypothetical protein POPTR_0015s04770g [Populus trichocarpa] |
| 4 |
Hb_000402_060 |
0.1072909347 |
- |
- |
PREDICTED: uncharacterized protein LOC105644642 isoform X2 [Jatropha curcas] |
| 5 |
Hb_158092_090 |
0.1179884922 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 6 |
Hb_000377_130 |
0.1195962733 |
- |
- |
hypothetical protein POPTR_0016s00790g [Populus trichocarpa] |
| 7 |
Hb_000620_050 |
0.1240189999 |
- |
- |
PREDICTED: uncharacterized protein LOC105649924 [Jatropha curcas] |
| 8 |
Hb_001408_200 |
0.1246126102 |
- |
- |
hypothetical protein JCGZ_08964 [Jatropha curcas] |
| 9 |
Hb_006755_030 |
0.1275502814 |
- |
- |
BURP domain-containing protein, putative [Theobroma cacao] |
| 10 |
Hb_002400_300 |
0.1286834697 |
- |
- |
PREDICTED: synaptotagmin-4 [Jatropha curcas] |
| 11 |
Hb_000122_230 |
0.1345635081 |
- |
- |
laccase, putative [Ricinus communis] |
| 12 |
Hb_002925_060 |
0.1357716435 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01547 [Jatropha curcas] |
| 13 |
Hb_000866_400 |
0.1366340837 |
- |
- |
- |
| 14 |
Hb_121624_010 |
0.1376871527 |
- |
- |
PREDICTED: uncharacterized protein LOC105634410 [Jatropha curcas] |
| 15 |
Hb_011501_020 |
0.1378618488 |
- |
- |
PREDICTED: uncharacterized protein LOC105648929 [Jatropha curcas] |
| 16 |
Hb_186765_010 |
0.1379286533 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105629453 [Jatropha curcas] |
| 17 |
Hb_025623_010 |
0.1384933146 |
- |
- |
- |
| 18 |
Hb_026317_040 |
0.1387160254 |
- |
- |
hypothetical protein POPTR_0009s12270g [Populus trichocarpa] |
| 19 |
Hb_003058_240 |
0.1405397879 |
- |
- |
PREDICTED: tubulin beta-5 chain-like [Jatropha curcas] |
| 20 |
Hb_002245_060 |
0.1407193321 |
- |
- |
PREDICTED: kinesin-3 isoform X1 [Jatropha curcas] |