| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
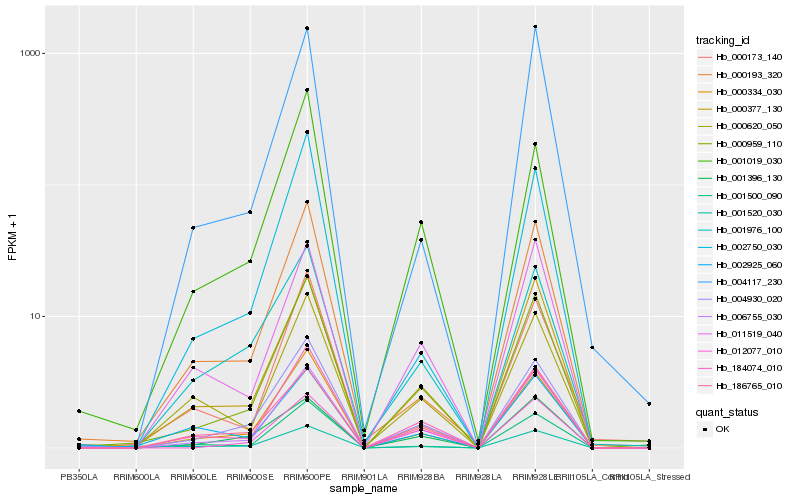
Hb_000959_110 |
0.0 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 13 [Vitis vinifera] |
| 2 |
Hb_006755_030 |
0.0696667254 |
- |
- |
BURP domain-containing protein, putative [Theobroma cacao] |
| 3 |
Hb_004930_020 |
0.0757933802 |
transcription factor |
TF Family: MYB |
PREDICTED: transcriptional activator Myb-like [Jatropha curcas] |
| 4 |
Hb_184074_010 |
0.0811055808 |
- |
- |
PREDICTED: uncharacterized protein LOC105639054 [Jatropha curcas] |
| 5 |
Hb_000334_030 |
0.0829573891 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000377_130 |
0.0841971391 |
- |
- |
hypothetical protein POPTR_0016s00790g [Populus trichocarpa] |
| 7 |
Hb_186765_010 |
0.1190284286 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105629453 [Jatropha curcas] |
| 8 |
Hb_011519_040 |
0.1198487478 |
- |
- |
hypothetical protein POPTR_0015s04770g [Populus trichocarpa] |
| 9 |
Hb_000620_050 |
0.1209608703 |
- |
- |
PREDICTED: uncharacterized protein LOC105649924 [Jatropha curcas] |
| 10 |
Hb_000193_320 |
0.1242878605 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001396_130 |
0.1255315792 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001500_090 |
0.1274075232 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001019_030 |
0.1281077127 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase 1-like [Jatropha curcas] |
| 14 |
Hb_001976_100 |
0.1283006619 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_012077_010 |
0.1290412952 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 16 |
Hb_002750_030 |
0.1341612683 |
- |
- |
PREDICTED: protein IRX15-LIKE-like [Populus euphratica] |
| 17 |
Hb_000173_140 |
0.1399954812 |
- |
- |
EF-hand calcium-binding domain-containing protein 4A [Theobroma cacao] |
| 18 |
Hb_004117_230 |
0.1402419613 |
- |
- |
lipid transfer precursor protein [Hevea brasiliensis] |
| 19 |
Hb_002925_060 |
0.1417498509 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01547 [Jatropha curcas] |
| 20 |
Hb_001520_030 |
0.1420682863 |
transcription factor |
TF Family: MYB |
PREDICTED: trichome differentiation protein GL1-like isoform X2 [Populus euphratica] |