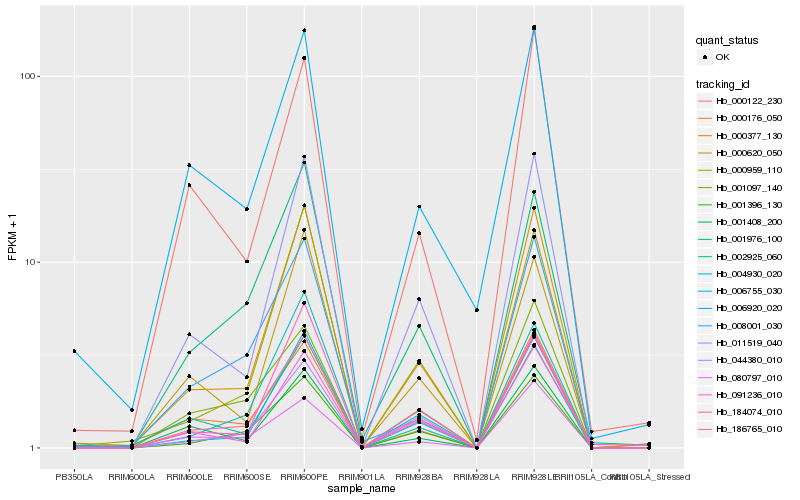
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_186765_010 |
0.0 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105629453 [Jatropha curcas] |
| 2 |
Hb_001396_130 |
0.0662694493 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000377_130 |
0.0665466871 |
- |
- |
hypothetical protein POPTR_0016s00790g [Populus trichocarpa] |
| 4 |
Hb_011519_040 |
0.0671345273 |
- |
- |
hypothetical protein POPTR_0015s04770g [Populus trichocarpa] |
| 5 |
Hb_006755_030 |
0.0854659615 |
- |
- |
BURP domain-containing protein, putative [Theobroma cacao] |
| 6 |
Hb_001097_140 |
0.0988640199 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002925_060 |
0.1000819076 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01547 [Jatropha curcas] |
| 8 |
Hb_044380_010 |
0.1046268212 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Malus domestica] |
| 9 |
Hb_080797_010 |
0.1053674255 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570-like [Citrus sinensis] |
| 10 |
Hb_001408_200 |
0.1080246039 |
- |
- |
hypothetical protein JCGZ_08964 [Jatropha curcas] |
| 11 |
Hb_001976_100 |
0.108369997 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000176_050 |
0.1121757256 |
- |
- |
glutamine amidotransferase class-I domain-containing family protein [Populus trichocarpa] |
| 13 |
Hb_091236_010 |
0.1152265315 |
- |
- |
hypothetical protein POPTR_0019s12290g [Populus trichocarpa] |
| 14 |
Hb_000122_230 |
0.1187963614 |
- |
- |
laccase, putative [Ricinus communis] |
| 15 |
Hb_000959_110 |
0.1190284286 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 13 [Vitis vinifera] |
| 16 |
Hb_000620_050 |
0.1201792707 |
- |
- |
PREDICTED: uncharacterized protein LOC105649924 [Jatropha curcas] |
| 17 |
Hb_004930_020 |
0.1238654326 |
transcription factor |
TF Family: MYB |
PREDICTED: transcriptional activator Myb-like [Jatropha curcas] |
| 18 |
Hb_008001_030 |
0.1254602138 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_184074_010 |
0.1259896617 |
- |
- |
PREDICTED: uncharacterized protein LOC105639054 [Jatropha curcas] |
| 20 |
Hb_006920_020 |
0.1262826395 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |