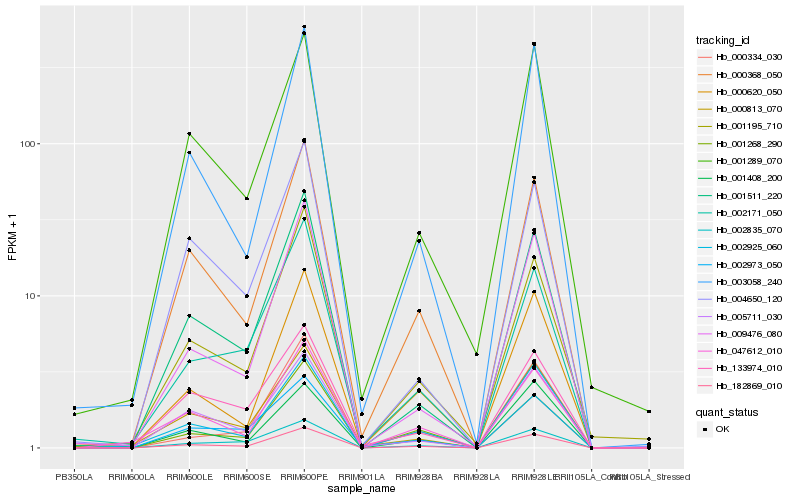
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_182869_010 |
0.0 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 2 |
Hb_000368_050 |
0.0586609341 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 3 |
Hb_001511_220 |
0.0755484553 |
- |
- |
hypothetical protein POPTR_0008s06990g [Populus trichocarpa] |
| 4 |
Hb_002925_060 |
0.0836901749 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01547 [Jatropha curcas] |
| 5 |
Hb_000620_050 |
0.0874799972 |
- |
- |
PREDICTED: uncharacterized protein LOC105649924 [Jatropha curcas] |
| 6 |
Hb_005711_030 |
0.0891413615 |
- |
- |
hypothetical protein RCOM_0460020 [Ricinus communis] |
| 7 |
Hb_003058_240 |
0.0922135128 |
- |
- |
PREDICTED: tubulin beta-5 chain-like [Jatropha curcas] |
| 8 |
Hb_000813_070 |
0.0929656242 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001195_710 |
0.0933353929 |
- |
- |
glycosyltransferase family GT8 protein [Populus deltoides] |
| 10 |
Hb_002835_070 |
0.0938587478 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_009476_080 |
0.0941689878 |
- |
- |
BnaA03g18920D [Brassica napus] |
| 12 |
Hb_001289_070 |
0.0956380926 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 13 |
Hb_047612_010 |
0.0965401787 |
- |
- |
glutamine amidotransferase class-I domain-containing family protein [Populus trichocarpa] |
| 14 |
Hb_001268_290 |
0.0979767806 |
- |
- |
PREDICTED: glutaredoxin-C1-like [Jatropha curcas] |
| 15 |
Hb_002973_050 |
0.0999999773 |
- |
- |
hypothetical protein JCGZ_06491 [Jatropha curcas] |
| 16 |
Hb_002171_050 |
0.1005624715 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase PEPR1 [Populus euphratica] |
| 17 |
Hb_133974_010 |
0.1058914148 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 3-like isoform X2 [Populus euphratica] |
| 18 |
Hb_000334_030 |
0.1064145315 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004650_120 |
0.1077965389 |
- |
- |
PREDICTED: auxin transporter-like protein 3 [Jatropha curcas] |
| 20 |
Hb_001408_200 |
0.1082216084 |
- |
- |
hypothetical protein JCGZ_08964 [Jatropha curcas] |