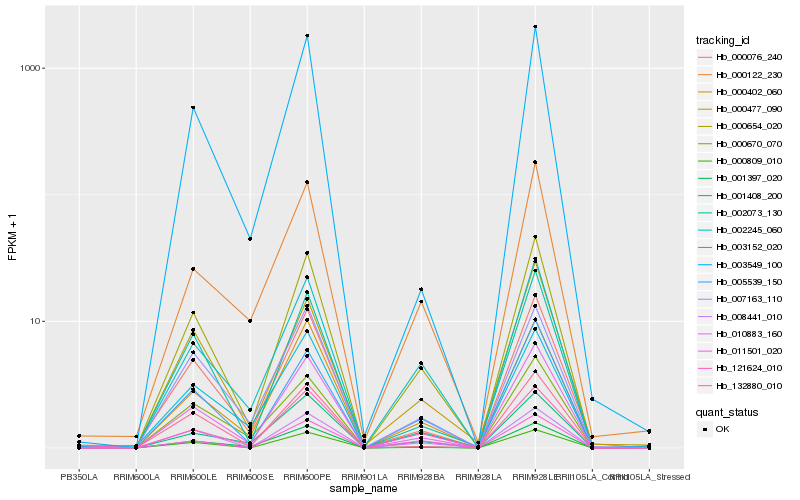
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000654_020 |
0.0 |
- |
- |
hypothetical protein CICLE_v10006262mg [Citrus clementina] |
| 2 |
Hb_010883_160 |
0.0527804354 |
- |
- |
PREDICTED: pathogenesis-related protein 5 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000809_010 |
0.0643922261 |
- |
- |
hypothetical protein JCGZ_12342 [Jatropha curcas] |
| 4 |
Hb_005539_150 |
0.0734788235 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 33 [Jatropha curcas] |
| 5 |
Hb_008441_010 |
0.0767217135 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 6 |
Hb_002245_060 |
0.0768608035 |
- |
- |
PREDICTED: kinesin-3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000402_060 |
0.0831265519 |
- |
- |
PREDICTED: uncharacterized protein LOC105644642 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000670_070 |
0.0847521892 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 9 |
Hb_002073_130 |
0.0889886699 |
- |
- |
PREDICTED: uncharacterized protein LOC100246830 [Vitis vinifera] |
| 10 |
Hb_000076_240 |
0.0896581613 |
- |
- |
Early nodulin 55-2 precursor, putative [Ricinus communis] |
| 11 |
Hb_001408_200 |
0.096514488 |
- |
- |
hypothetical protein JCGZ_08964 [Jatropha curcas] |
| 12 |
Hb_000477_090 |
0.1017805925 |
- |
- |
PREDICTED: uncharacterized protein LOC105639302 [Jatropha curcas] |
| 13 |
Hb_001397_020 |
0.1020193982 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 14 |
Hb_007163_110 |
0.1033120861 |
- |
- |
PREDICTED: kinesin-like protein KIF19 [Jatropha curcas] |
| 15 |
Hb_132880_010 |
0.1065466645 |
- |
- |
PREDICTED: uncharacterized protein LOC105641595 [Jatropha curcas] |
| 16 |
Hb_000122_230 |
0.1066379353 |
- |
- |
laccase, putative [Ricinus communis] |
| 17 |
Hb_003152_020 |
0.1068045634 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Jatropha curcas] |
| 18 |
Hb_011501_020 |
0.1071174631 |
- |
- |
PREDICTED: uncharacterized protein LOC105648929 [Jatropha curcas] |
| 19 |
Hb_003549_100 |
0.1100937752 |
- |
- |
PREDICTED: uncharacterized protein LOC105647410 [Jatropha curcas] |
| 20 |
Hb_121624_010 |
0.1104232181 |
- |
- |
PREDICTED: uncharacterized protein LOC105634410 [Jatropha curcas] |