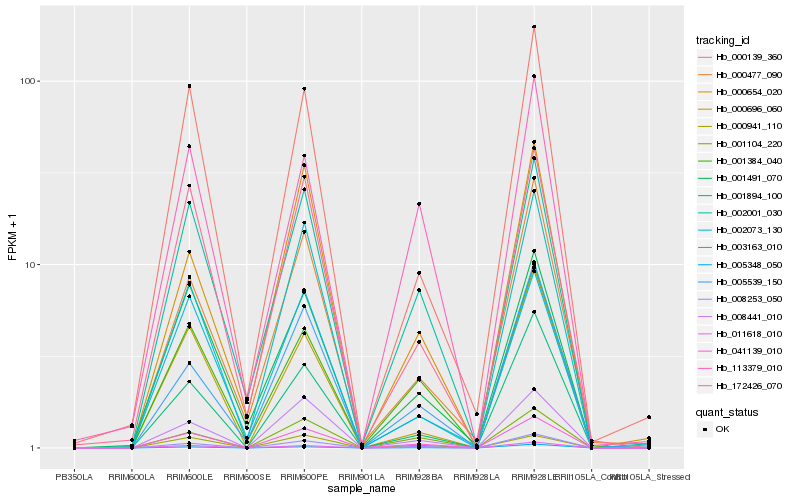
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_041139_010 |
0.0 |
- |
- |
PREDICTED: probable terpene synthase 13 [Jatropha curcas] |
| 2 |
Hb_008253_050 |
0.0546574805 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0001s36220g [Populus trichocarpa] |
| 3 |
Hb_011618_010 |
0.0648819422 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g59700 [Jatropha curcas] |
| 4 |
Hb_008441_010 |
0.0876506362 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 5 |
Hb_002001_030 |
0.0941423962 |
- |
- |
Plasma membrane ATPase 4 [Gossypium arboreum] |
| 6 |
Hb_001894_100 |
0.0974399274 |
- |
- |
PREDICTED: kininogen-1-like [Jatropha curcas] |
| 7 |
Hb_172426_070 |
0.0975414114 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 3-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000139_360 |
0.0984721552 |
- |
- |
PREDICTED: uncharacterized protein LOC105631933 [Jatropha curcas] |
| 9 |
Hb_001104_220 |
0.0990401204 |
- |
- |
PREDICTED: uncharacterized protein LOC105629453 [Jatropha curcas] |
| 10 |
Hb_001384_040 |
0.1010732255 |
- |
- |
PREDICTED: beta-glucosidase 17-like [Jatropha curcas] |
| 11 |
Hb_001491_070 |
0.1086789613 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 17 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003163_010 |
0.1153161729 |
- |
- |
hypothetical protein POPTR_0012s02920g [Populus trichocarpa] |
| 13 |
Hb_000477_090 |
0.1192832059 |
- |
- |
PREDICTED: uncharacterized protein LOC105639302 [Jatropha curcas] |
| 14 |
Hb_005348_050 |
0.1218969356 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 15 |
Hb_005539_150 |
0.1255199052 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 33 [Jatropha curcas] |
| 16 |
Hb_113379_010 |
0.1257014375 |
- |
- |
PREDICTED: beta-xylosidase/alpha-L-arabinofuranosidase 1 [Jatropha curcas] |
| 17 |
Hb_000941_110 |
0.1289070869 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 18 |
Hb_000696_060 |
0.1313479029 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000654_020 |
0.1318899145 |
- |
- |
hypothetical protein CICLE_v10006262mg [Citrus clementina] |
| 20 |
Hb_002073_130 |
0.1324239624 |
- |
- |
PREDICTED: uncharacterized protein LOC100246830 [Vitis vinifera] |