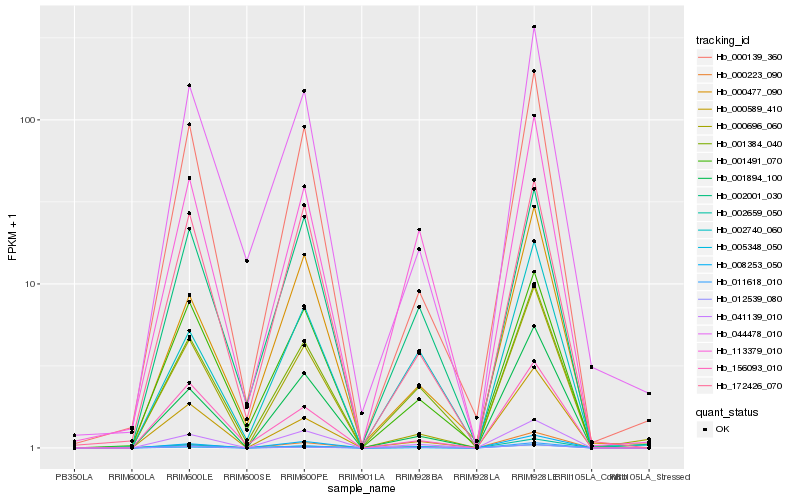
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011618_010 |
0.0 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g59700 [Jatropha curcas] |
| 2 |
Hb_041139_010 |
0.0648819422 |
- |
- |
PREDICTED: probable terpene synthase 13 [Jatropha curcas] |
| 3 |
Hb_008253_050 |
0.069223414 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0001s36220g [Populus trichocarpa] |
| 4 |
Hb_001384_040 |
0.0741980564 |
- |
- |
PREDICTED: beta-glucosidase 17-like [Jatropha curcas] |
| 5 |
Hb_005348_050 |
0.0815620845 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 6 |
Hb_001894_100 |
0.0881310495 |
- |
- |
PREDICTED: kininogen-1-like [Jatropha curcas] |
| 7 |
Hb_000589_410 |
0.0882272534 |
- |
- |
PREDICTED: thylakoid lumenal 15 kDa protein 1, chloroplastic-like isoform X1 [Populus euphratica] |
| 8 |
Hb_000139_360 |
0.0918231766 |
- |
- |
PREDICTED: uncharacterized protein LOC105631933 [Jatropha curcas] |
| 9 |
Hb_113379_010 |
0.0990381985 |
- |
- |
PREDICTED: beta-xylosidase/alpha-L-arabinofuranosidase 1 [Jatropha curcas] |
| 10 |
Hb_002659_050 |
0.1091871431 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 10 isoform X3 [Jatropha curcas] |
| 11 |
Hb_000696_060 |
0.1098823906 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000223_090 |
0.1104433543 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 13 |
Hb_001491_070 |
0.1171934847 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 17 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002001_030 |
0.1233366244 |
- |
- |
Plasma membrane ATPase 4 [Gossypium arboreum] |
| 15 |
Hb_002740_060 |
0.1249745032 |
- |
- |
PREDICTED: pectin acetylesterase 12 [Jatropha curcas] |
| 16 |
Hb_044478_010 |
0.1273255078 |
- |
- |
plastoquinol-plastocyanin reductase, putative [Ricinus communis] |
| 17 |
Hb_156093_010 |
0.1280429383 |
- |
- |
hypothetical protein RCOM_0504250 [Ricinus communis] |
| 18 |
Hb_172426_070 |
0.129060575 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 3-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_012539_080 |
0.1306245825 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 20 |
Hb_000477_090 |
0.1310982626 |
- |
- |
PREDICTED: uncharacterized protein LOC105639302 [Jatropha curcas] |