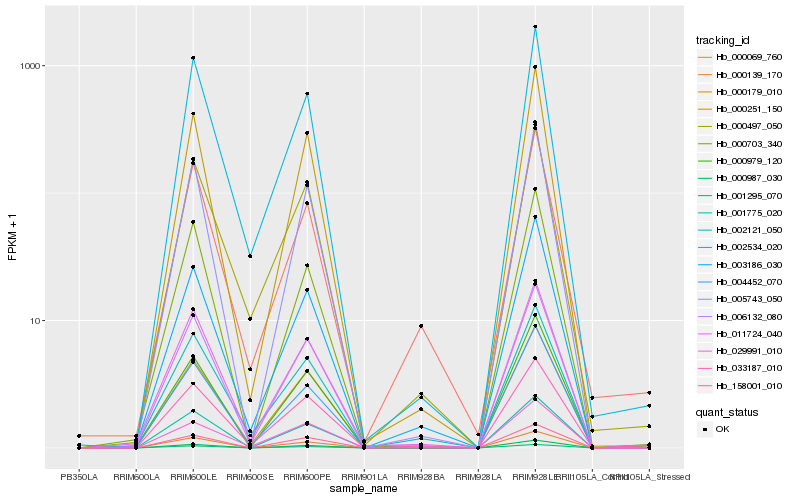
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006132_080 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 86A8-like [Jatropha curcas] |
| 2 |
Hb_002121_050 |
0.0534257025 |
- |
- |
hypothetical protein EUGRSUZ_D02586 [Eucalyptus grandis] |
| 3 |
Hb_003186_030 |
0.0535350574 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0012s05720g [Populus trichocarpa] |
| 4 |
Hb_004452_070 |
0.0544634881 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 5 |
Hb_000251_150 |
0.0600211391 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 6 |
Hb_000179_010 |
0.0601339608 |
- |
- |
PREDICTED: alkane hydroxylase MAH1-like [Jatropha curcas] |
| 7 |
Hb_011724_040 |
0.0627867674 |
- |
- |
PREDICTED: uncharacterized protein LOC105632860 [Jatropha curcas] |
| 8 |
Hb_005743_050 |
0.0660318503 |
- |
- |
unnamed protein product [Coffea canephora] |
| 9 |
Hb_033187_010 |
0.0696344607 |
- |
- |
epoxide hydrolase, putative [Ricinus communis] |
| 10 |
Hb_002534_020 |
0.0713983422 |
- |
- |
PREDICTED: chlorophyll a-b binding protein 4, chloroplastic [Gossypium raimondii] |
| 11 |
Hb_000139_170 |
0.0718534956 |
- |
- |
PREDICTED: uncharacterized protein LOC105631957 [Jatropha curcas] |
| 12 |
Hb_000987_030 |
0.0722713606 |
transcription factor |
TF Family: BES1 |
hypothetical protein POPTR_0016s13350g [Populus trichocarpa] |
| 13 |
Hb_000497_050 |
0.0737557809 |
- |
- |
PREDICTED: photosystem II repair protein PSB27-H1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000979_120 |
0.0747327822 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 15 |
Hb_001775_020 |
0.0755987073 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 16 |
Hb_001295_070 |
0.0757417424 |
- |
- |
PREDICTED: putative UPF0481 protein At3g02645 [Jatropha curcas] |
| 17 |
Hb_158001_010 |
0.0774592038 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1, partial [Populus euphratica] |
| 18 |
Hb_029991_010 |
0.0778295128 |
- |
- |
hypothetical protein PRUPE_ppa001615mg [Prunus persica] |
| 19 |
Hb_000069_760 |
0.0781055042 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1A, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000703_340 |
0.0781427631 |
- |
- |
Oxygen-evolving enhancer protein 2, chloroplast precursor, putative [Ricinus communis] |