| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
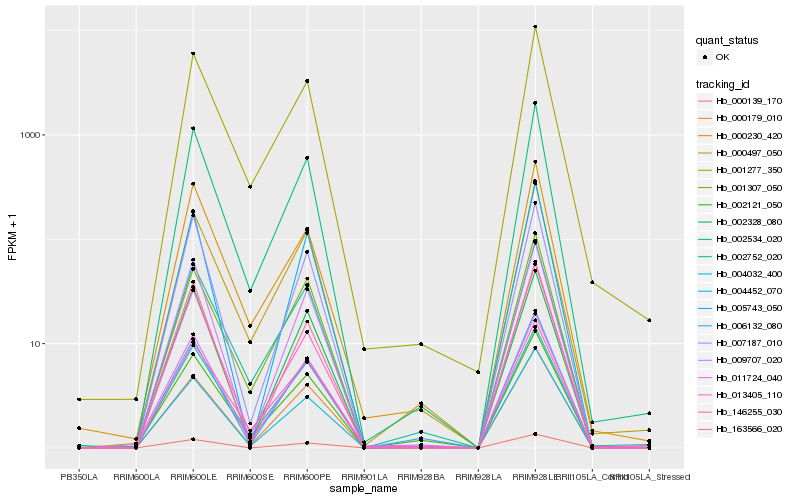
Hb_002534_020 |
0.0 |
- |
- |
PREDICTED: chlorophyll a-b binding protein 4, chloroplastic [Gossypium raimondii] |
| 2 |
Hb_001277_350 |
0.0425252676 |
- |
- |
early light-induced protein, putative [Ricinus communis] |
| 3 |
Hb_146255_030 |
0.0500537508 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HY5-like [Jatropha curcas] |
| 4 |
Hb_000497_050 |
0.0502989358 |
- |
- |
PREDICTED: photosystem II repair protein PSB27-H1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_002752_020 |
0.0523237343 |
- |
- |
FKBP-type peptidyl-prolyl cis-trans isomerase 3 family protein [Populus trichocarpa] |
| 6 |
Hb_011724_040 |
0.0569251542 |
- |
- |
PREDICTED: uncharacterized protein LOC105632860 [Jatropha curcas] |
| 7 |
Hb_004032_400 |
0.057807504 |
- |
- |
PREDICTED: uncharacterized transporter YBR287W-like [Jatropha curcas] |
| 8 |
Hb_004452_070 |
0.0594371341 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 9 |
Hb_000230_420 |
0.0594509393 |
- |
- |
GS [Hevea brasiliensis subsp. brasiliensis] |
| 10 |
Hb_000179_010 |
0.0633278107 |
- |
- |
PREDICTED: alkane hydroxylase MAH1-like [Jatropha curcas] |
| 11 |
Hb_013405_110 |
0.0636646957 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 12 |
Hb_001307_050 |
0.0638725917 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.2-like [Jatropha curcas] |
| 13 |
Hb_009707_020 |
0.0658862784 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 12 [Jatropha curcas] |
| 14 |
Hb_002121_050 |
0.0661089306 |
- |
- |
hypothetical protein EUGRSUZ_D02586 [Eucalyptus grandis] |
| 15 |
Hb_000139_170 |
0.0684991903 |
- |
- |
PREDICTED: uncharacterized protein LOC105631957 [Jatropha curcas] |
| 16 |
Hb_005743_050 |
0.0705566211 |
- |
- |
unnamed protein product [Coffea canephora] |
| 17 |
Hb_163566_020 |
0.0708614102 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP17-1, chloroplastic isoform X2 [Jatropha curcas] |
| 18 |
Hb_007187_010 |
0.0709791013 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 19 |
Hb_006132_080 |
0.0713983422 |
- |
- |
PREDICTED: cytochrome P450 86A8-like [Jatropha curcas] |
| 20 |
Hb_002328_080 |
0.0714023423 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |