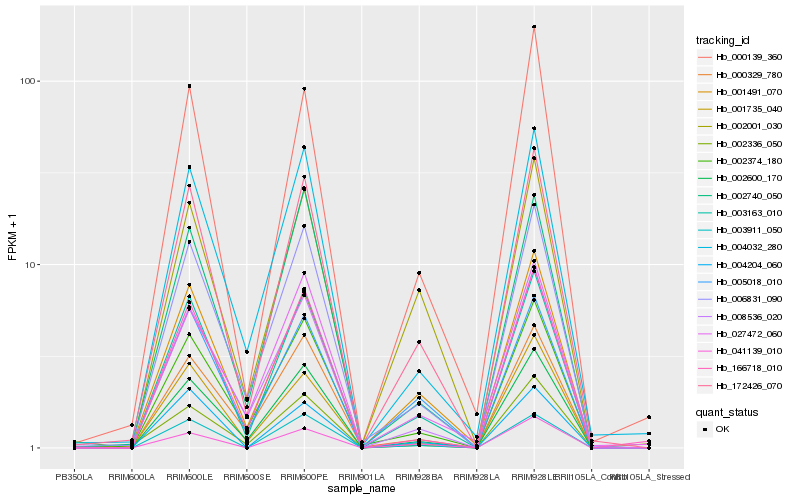
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_172426_070 |
0.0 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 3-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_003163_010 |
0.0447706375 |
- |
- |
hypothetical protein POPTR_0012s02920g [Populus trichocarpa] |
| 3 |
Hb_006831_090 |
0.0685300374 |
- |
- |
PREDICTED: uncharacterized protein LOC105642125 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001491_070 |
0.0694311708 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 17 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002600_170 |
0.0822813847 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004032_280 |
0.0834516997 |
- |
- |
PREDICTED: uncharacterized protein LOC105635149 [Jatropha curcas] |
| 7 |
Hb_008536_020 |
0.0893417795 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 8 |
Hb_000139_360 |
0.0893730414 |
- |
- |
PREDICTED: uncharacterized protein LOC105631933 [Jatropha curcas] |
| 9 |
Hb_003911_050 |
0.0933601476 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |
| 10 |
Hb_005018_010 |
0.0937192534 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 11 |
Hb_002001_030 |
0.0964254049 |
- |
- |
Plasma membrane ATPase 4 [Gossypium arboreum] |
| 12 |
Hb_002374_180 |
0.0971676228 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-related [Jatropha curcas] |
| 13 |
Hb_041139_010 |
0.0975414114 |
- |
- |
PREDICTED: probable terpene synthase 13 [Jatropha curcas] |
| 14 |
Hb_001735_040 |
0.0981241377 |
- |
- |
PREDICTED: F-box protein SKIP2-like isoform X2 [Populus euphratica] |
| 15 |
Hb_000329_780 |
0.1033805619 |
- |
- |
hypothetical protein POPTR_0403s00200g [Populus trichocarpa] |
| 16 |
Hb_002336_050 |
0.1034541111 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_166718_010 |
0.1035336313 |
- |
- |
heat-shock protein, putative [Ricinus communis] |
| 18 |
Hb_002740_050 |
0.1054178496 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86-like [Populus euphratica] |
| 19 |
Hb_027472_060 |
0.106691618 |
transcription factor |
TF Family: GRAS |
DELLA protein GAI1, putative [Ricinus communis] |
| 20 |
Hb_004204_060 |
0.1074712175 |
transcription factor |
TF Family: NAC |
NAC transcription factor 058, partial [Jatropha curcas] |