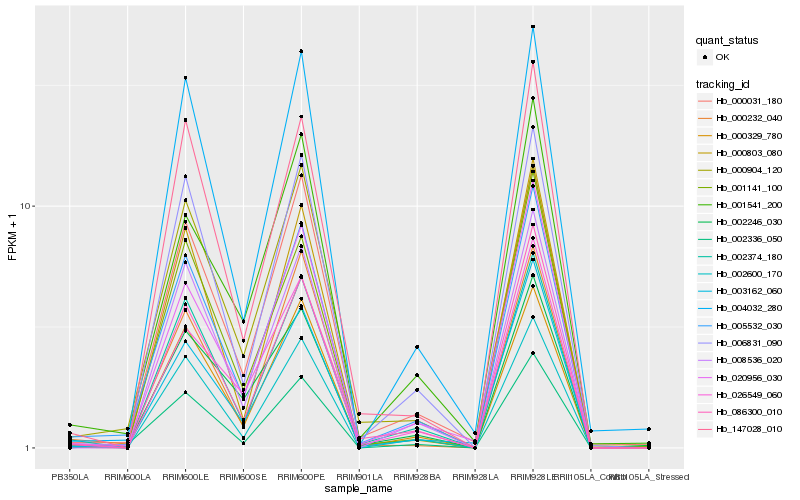
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005532_030 |
0.0 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_002374_180 |
0.0663123721 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-related [Jatropha curcas] |
| 3 |
Hb_002336_050 |
0.0702372471 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_002600_170 |
0.0704248648 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_006831_090 |
0.0712226442 |
- |
- |
PREDICTED: uncharacterized protein LOC105642125 isoform X1 [Jatropha curcas] |
| 6 |
Hb_086300_010 |
0.0750571334 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 7 |
Hb_000329_780 |
0.0779104169 |
- |
- |
hypothetical protein POPTR_0403s00200g [Populus trichocarpa] |
| 8 |
Hb_000031_180 |
0.0794930721 |
- |
- |
sarcosine oxidase, putative [Ricinus communis] |
| 9 |
Hb_020956_030 |
0.0809293276 |
- |
- |
PREDICTED: swi5-dependent recombination DNA repair protein 1 homolog [Jatropha curcas] |
| 10 |
Hb_008536_020 |
0.0813731492 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 11 |
Hb_003162_060 |
0.0813825961 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 12 |
Hb_001141_100 |
0.0841997514 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2-like [Jatropha curcas] |
| 13 |
Hb_147028_010 |
0.0861449258 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HD1 [Prunus mume] |
| 14 |
Hb_002246_030 |
0.0871505713 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 15 |
Hb_004032_280 |
0.0873634393 |
- |
- |
PREDICTED: uncharacterized protein LOC105635149 [Jatropha curcas] |
| 16 |
Hb_026549_060 |
0.0935795522 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 17 |
Hb_001541_200 |
0.0939864046 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |
| 18 |
Hb_000904_120 |
0.0942574607 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000232_040 |
0.0949800722 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 86 isoform X2 [Populus euphratica] |
| 20 |
Hb_000803_080 |
0.0967563105 |
desease resistance |
Gene Name: ABC_tran |
ATP-binding cassette transporter, putative [Ricinus communis] |