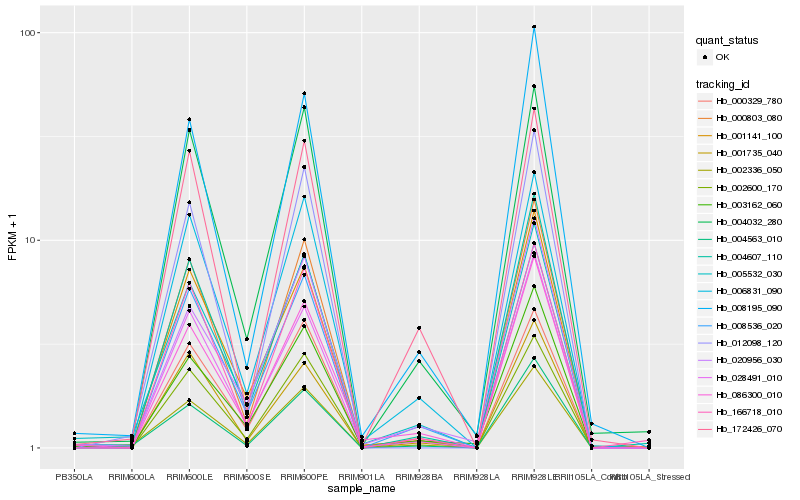
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002336_050 |
0.0 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_004563_010 |
0.0603037116 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_005532_030 |
0.0702372471 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_008536_020 |
0.0851313158 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 5 |
Hb_006831_090 |
0.0871053453 |
- |
- |
PREDICTED: uncharacterized protein LOC105642125 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002600_170 |
0.0909429072 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004032_280 |
0.0924561445 |
- |
- |
PREDICTED: uncharacterized protein LOC105635149 [Jatropha curcas] |
| 8 |
Hb_012098_120 |
0.0958698921 |
- |
- |
PREDICTED: putative elongation of fatty acids protein DDB_G0272012 [Jatropha curcas] |
| 9 |
Hb_000803_080 |
0.0970163002 |
desease resistance |
Gene Name: ABC_tran |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 10 |
Hb_028491_010 |
0.0988884292 |
- |
- |
hypothetical protein JCGZ_12857 [Jatropha curcas] |
| 11 |
Hb_166718_010 |
0.0998250946 |
- |
- |
heat-shock protein, putative [Ricinus communis] |
| 12 |
Hb_000329_780 |
0.1014795243 |
- |
- |
hypothetical protein POPTR_0403s00200g [Populus trichocarpa] |
| 13 |
Hb_001141_100 |
0.1016477246 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2-like [Jatropha curcas] |
| 14 |
Hb_004607_110 |
0.1024212578 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek6 [Jatropha curcas] |
| 15 |
Hb_003162_060 |
0.1032722719 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 16 |
Hb_172426_070 |
0.1034541111 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 3-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_008195_090 |
0.1049872528 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_086300_010 |
0.1062018048 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 19 |
Hb_001735_040 |
0.1074278012 |
- |
- |
PREDICTED: F-box protein SKIP2-like isoform X2 [Populus euphratica] |
| 20 |
Hb_020956_030 |
0.1077076583 |
- |
- |
PREDICTED: swi5-dependent recombination DNA repair protein 1 homolog [Jatropha curcas] |