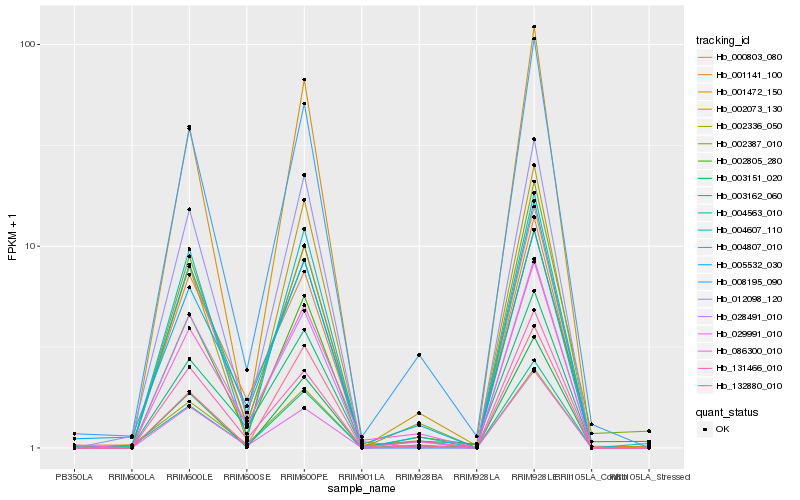
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004563_010 |
0.0 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_002336_050 |
0.0603037116 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_008195_090 |
0.0708605331 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_012098_120 |
0.0798405228 |
- |
- |
PREDICTED: putative elongation of fatty acids protein DDB_G0272012 [Jatropha curcas] |
| 5 |
Hb_003151_020 |
0.082468307 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 6 |
Hb_029991_010 |
0.0894436769 |
- |
- |
hypothetical protein PRUPE_ppa001615mg [Prunus persica] |
| 7 |
Hb_004607_110 |
0.0899499683 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek6 [Jatropha curcas] |
| 8 |
Hb_000803_080 |
0.0920869663 |
desease resistance |
Gene Name: ABC_tran |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 9 |
Hb_028491_010 |
0.0937408894 |
- |
- |
hypothetical protein JCGZ_12857 [Jatropha curcas] |
| 10 |
Hb_003162_060 |
0.0987222956 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 11 |
Hb_001472_150 |
0.0989039005 |
- |
- |
PREDICTED: cytochrome P450 86A7 [Jatropha curcas] |
| 12 |
Hb_005532_030 |
0.0994232341 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_002073_130 |
0.1002940187 |
- |
- |
PREDICTED: uncharacterized protein LOC100246830 [Vitis vinifera] |
| 14 |
Hb_002805_280 |
0.1012498042 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 15 |
Hb_131466_010 |
0.1016950455 |
- |
- |
PREDICTED: putative amidase C869.01 [Jatropha curcas] |
| 16 |
Hb_132880_010 |
0.1024551325 |
- |
- |
PREDICTED: uncharacterized protein LOC105641595 [Jatropha curcas] |
| 17 |
Hb_002387_010 |
0.1025364664 |
- |
- |
PREDICTED: protein STAY-GREEN LIKE, chloroplastic [Jatropha curcas] |
| 18 |
Hb_086300_010 |
0.1034135701 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 19 |
Hb_004807_010 |
0.1038859203 |
- |
- |
PREDICTED: auxin-induced protein 15A-like [Jatropha curcas] |
| 20 |
Hb_001141_100 |
0.1045765853 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2-like [Jatropha curcas] |