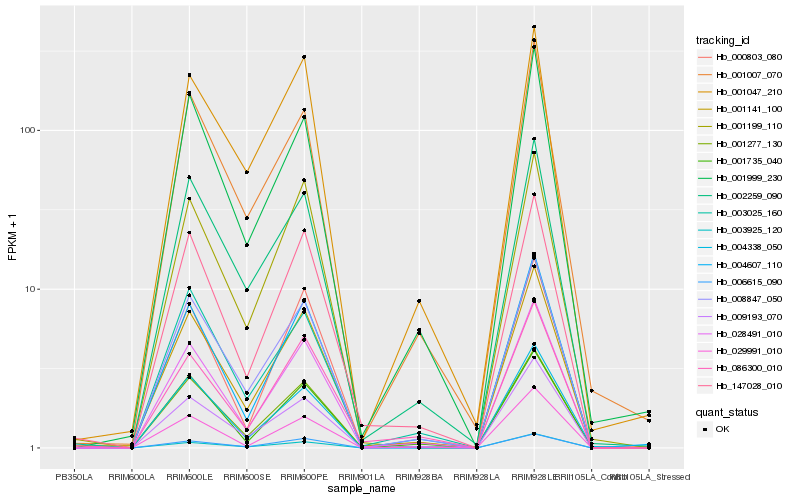
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001141_100 |
0.0 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2-like [Jatropha curcas] |
| 2 |
Hb_028491_010 |
0.0460586455 |
- |
- |
hypothetical protein JCGZ_12857 [Jatropha curcas] |
| 3 |
Hb_008847_050 |
0.0554676737 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_147028_010 |
0.0556609904 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HD1 [Prunus mume] |
| 5 |
Hb_004607_110 |
0.0590380054 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek6 [Jatropha curcas] |
| 6 |
Hb_001277_130 |
0.0607530302 |
- |
- |
PREDICTED: dentin sialophosphoprotein-like [Jatropha curcas] |
| 7 |
Hb_009193_070 |
0.0653050864 |
- |
- |
fatty acid elongase 3-ketoacyl-CoA synthase 1 [Arabidopsis thaliana] |
| 8 |
Hb_002259_090 |
0.0723086928 |
- |
- |
PREDICTED: ABC transporter G family member 11 [Jatropha curcas] |
| 9 |
Hb_001199_110 |
0.0741911043 |
- |
- |
PREDICTED: uncharacterized protein ycf36 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001999_230 |
0.0754000354 |
- |
- |
PREDICTED: uncharacterized protein LOC105630678 [Jatropha curcas] |
| 11 |
Hb_003025_160 |
0.0758804092 |
- |
- |
Protein HVA22, putative [Ricinus communis] |
| 12 |
Hb_000803_080 |
0.0762622903 |
desease resistance |
Gene Name: ABC_tran |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 13 |
Hb_001007_070 |
0.0787926186 |
- |
- |
thioredoxin m(mitochondrial)-type, putative [Ricinus communis] |
| 14 |
Hb_006615_090 |
0.0791183334 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 15 |
Hb_004338_050 |
0.079627898 |
- |
- |
hypothetical protein POPTR_0004s02760g [Populus trichocarpa] |
| 16 |
Hb_086300_010 |
0.0812204461 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 17 |
Hb_001047_210 |
0.0821475111 |
- |
- |
PSI reaction center subunit II [Citrus sinensis] |
| 18 |
Hb_001735_040 |
0.0828810528 |
- |
- |
PREDICTED: F-box protein SKIP2-like isoform X2 [Populus euphratica] |
| 19 |
Hb_003925_120 |
0.0832662053 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 20 |
Hb_029991_010 |
0.0841092126 |
- |
- |
hypothetical protein PRUPE_ppa001615mg [Prunus persica] |