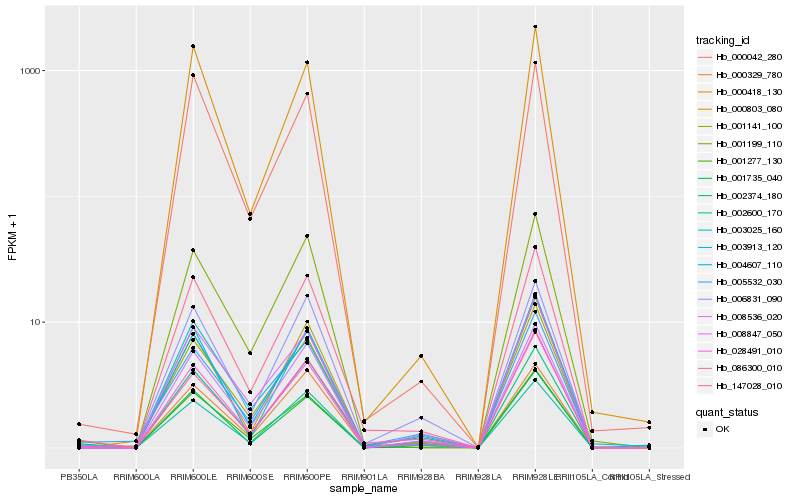
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_147028_010 |
0.0 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HD1 [Prunus mume] |
| 2 |
Hb_001141_100 |
0.0556609904 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2-like [Jatropha curcas] |
| 3 |
Hb_028491_010 |
0.0572833396 |
- |
- |
hypothetical protein JCGZ_12857 [Jatropha curcas] |
| 4 |
Hb_000803_080 |
0.0659922802 |
desease resistance |
Gene Name: ABC_tran |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 5 |
Hb_001277_130 |
0.0675906137 |
- |
- |
PREDICTED: dentin sialophosphoprotein-like [Jatropha curcas] |
| 6 |
Hb_086300_010 |
0.0708962855 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 7 |
Hb_001735_040 |
0.075287642 |
- |
- |
PREDICTED: F-box protein SKIP2-like isoform X2 [Populus euphratica] |
| 8 |
Hb_001199_110 |
0.0762509349 |
- |
- |
PREDICTED: uncharacterized protein ycf36 isoform X1 [Jatropha curcas] |
| 9 |
Hb_006831_090 |
0.0762849276 |
- |
- |
PREDICTED: uncharacterized protein LOC105642125 isoform X1 [Jatropha curcas] |
| 10 |
Hb_008536_020 |
0.0762918125 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 11 |
Hb_002374_180 |
0.076901975 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-related [Jatropha curcas] |
| 12 |
Hb_008847_050 |
0.0771791123 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004607_110 |
0.0787946758 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek6 [Jatropha curcas] |
| 14 |
Hb_002600_170 |
0.0828807563 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003025_160 |
0.0836644751 |
- |
- |
Protein HVA22, putative [Ricinus communis] |
| 16 |
Hb_000418_130 |
0.0839118954 |
- |
- |
PREDICTED: photosystem I reaction center subunit psaK, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000329_780 |
0.0860097294 |
- |
- |
hypothetical protein POPTR_0403s00200g [Populus trichocarpa] |
| 18 |
Hb_005532_030 |
0.0861449258 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_003913_120 |
0.0884904152 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s09430g [Populus trichocarpa] |
| 20 |
Hb_000042_280 |
0.0890348043 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |