| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
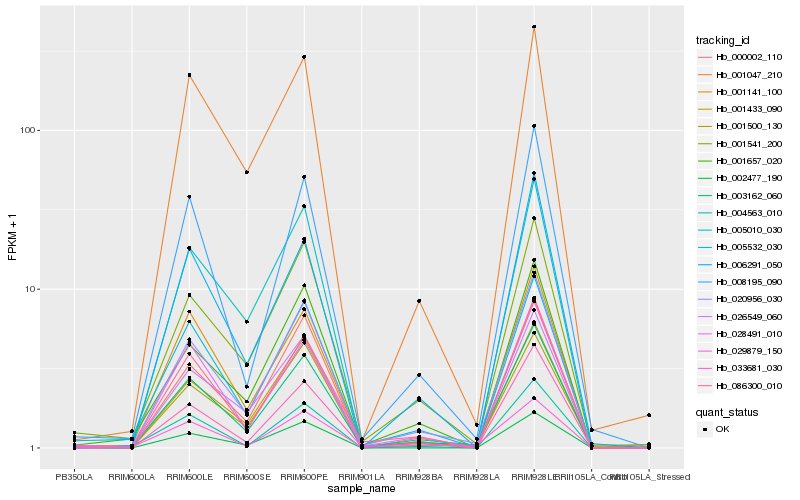
Hb_003162_060 |
0.0 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 2 |
Hb_020956_030 |
0.0316082777 |
- |
- |
PREDICTED: swi5-dependent recombination DNA repair protein 1 homolog [Jatropha curcas] |
| 3 |
Hb_086300_010 |
0.0733053838 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 4 |
Hb_005532_030 |
0.0813825961 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_026549_060 |
0.0843146415 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 6 |
Hb_001541_200 |
0.0852937746 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |
| 7 |
Hb_001141_100 |
0.0875687608 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2-like [Jatropha curcas] |
| 8 |
Hb_008195_090 |
0.0880551843 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001433_090 |
0.0887332216 |
- |
- |
Pectinesterase PPE8B precursor, putative [Ricinus communis] |
| 10 |
Hb_028491_010 |
0.0915209418 |
- |
- |
hypothetical protein JCGZ_12857 [Jatropha curcas] |
| 11 |
Hb_033681_030 |
0.0922249038 |
desease resistance |
Gene Name: NB-ARC |
Leucine-rich repeat containing protein [Theobroma cacao] |
| 12 |
Hb_005010_030 |
0.0941784214 |
- |
- |
PREDICTED: lecithin-cholesterol acyltransferase-like 1 [Jatropha curcas] |
| 13 |
Hb_001657_020 |
0.0953972929 |
- |
- |
heat shock family protein [Populus trichocarpa] |
| 14 |
Hb_001047_210 |
0.0963948171 |
- |
- |
PSI reaction center subunit II [Citrus sinensis] |
| 15 |
Hb_001500_130 |
0.0982356028 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |
| 16 |
Hb_004563_010 |
0.0987222956 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_002477_190 |
0.0997438778 |
- |
- |
hypothetical protein EUGRSUZ_C03380, partial [Eucalyptus grandis] |
| 18 |
Hb_006291_050 |
0.1001271603 |
- |
- |
PREDICTED: polyribonucleotide nucleotidyltransferase 2, mitochondrial isoform X1 [Jatropha curcas] |
| 19 |
Hb_029879_150 |
0.1004459829 |
desease resistance |
Gene Name: NB-ARC |
putative disease resistance gene NBS-LRR family protein [Populus trichocarpa] |
| 20 |
Hb_000002_110 |
0.1006370711 |
- |
- |
hypothetical protein EUGRSUZ_B02140 [Eucalyptus grandis] |