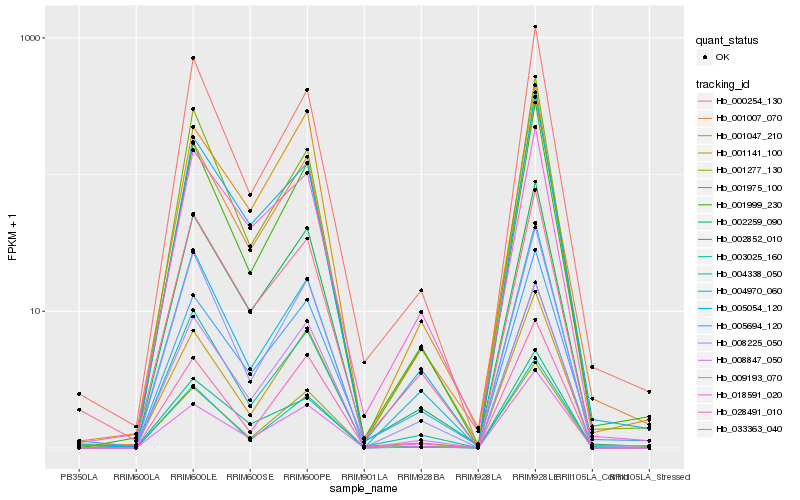
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002259_090 |
0.0 |
- |
- |
PREDICTED: ABC transporter G family member 11 [Jatropha curcas] |
| 2 |
Hb_008847_050 |
0.0395584907 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001007_070 |
0.0535916729 |
- |
- |
thioredoxin m(mitochondrial)-type, putative [Ricinus communis] |
| 4 |
Hb_003025_160 |
0.0563942675 |
- |
- |
Protein HVA22, putative [Ricinus communis] |
| 5 |
Hb_001999_230 |
0.0634248267 |
- |
- |
PREDICTED: uncharacterized protein LOC105630678 [Jatropha curcas] |
| 6 |
Hb_005054_120 |
0.0683811065 |
- |
- |
PREDICTED: sedoheptulose-1,7-bisphosphatase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001277_130 |
0.0685869503 |
- |
- |
PREDICTED: dentin sialophosphoprotein-like [Jatropha curcas] |
| 8 |
Hb_008225_050 |
0.0687027896 |
- |
- |
hypothetical protein PRUPE_ppa004332mg [Prunus persica] |
| 9 |
Hb_000254_130 |
0.0688448361 |
- |
- |
Photosystem I reaction center subunit VI, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_009193_070 |
0.0699680778 |
- |
- |
fatty acid elongase 3-ketoacyl-CoA synthase 1 [Arabidopsis thaliana] |
| 11 |
Hb_002852_010 |
0.0712525454 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 12 |
Hb_001141_100 |
0.0723086928 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2-like [Jatropha curcas] |
| 13 |
Hb_033363_040 |
0.0776968183 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 3 [Jatropha curcas] |
| 14 |
Hb_004970_060 |
0.0790763698 |
- |
- |
PREDICTED: uncharacterized protein LOC100259061 [Vitis vinifera] |
| 15 |
Hb_018591_020 |
0.0807209977 |
- |
- |
Ribulose bisphosphate carboxylase/oxygenase activase 1, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_001047_210 |
0.0812585793 |
- |
- |
PSI reaction center subunit II [Citrus sinensis] |
| 17 |
Hb_004338_050 |
0.0813474365 |
- |
- |
hypothetical protein POPTR_0004s02760g [Populus trichocarpa] |
| 18 |
Hb_028491_010 |
0.0821942072 |
- |
- |
hypothetical protein JCGZ_12857 [Jatropha curcas] |
| 19 |
Hb_005694_120 |
0.0843931335 |
- |
- |
PREDICTED: calcium-transporting ATPase, endoplasmic reticulum-type [Jatropha curcas] |
| 20 |
Hb_001975_100 |
0.0845324886 |
- |
- |
chlorophyll a-b binding protein 151, chloroplastic [Jatropha curcas] |