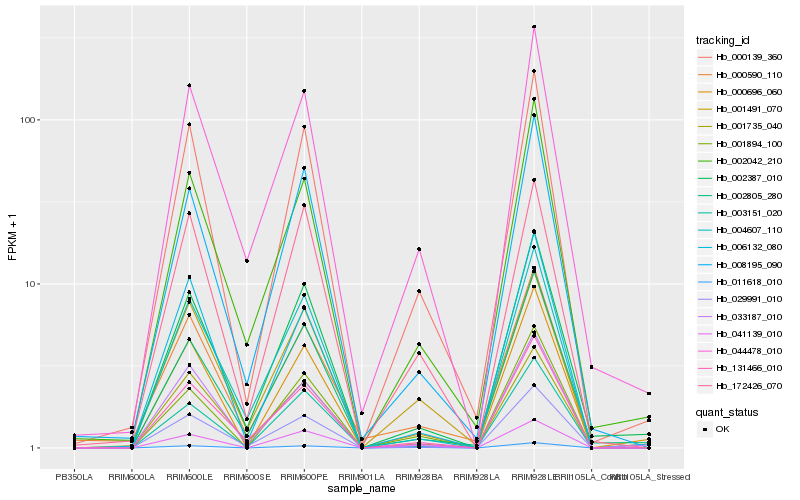
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000139_360 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631933 [Jatropha curcas] |
| 2 |
Hb_000696_060 |
0.0791771381 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_008195_090 |
0.0822819935 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_044478_010 |
0.0876685487 |
- |
- |
plastoquinol-plastocyanin reductase, putative [Ricinus communis] |
| 5 |
Hb_172426_070 |
0.0893730414 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 3-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_029991_010 |
0.0904977959 |
- |
- |
hypothetical protein PRUPE_ppa001615mg [Prunus persica] |
| 7 |
Hb_002387_010 |
0.0911206374 |
- |
- |
PREDICTED: protein STAY-GREEN LIKE, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001491_070 |
0.0912005888 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 17 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001735_040 |
0.0915401942 |
- |
- |
PREDICTED: F-box protein SKIP2-like isoform X2 [Populus euphratica] |
| 10 |
Hb_011618_010 |
0.0918231766 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g59700 [Jatropha curcas] |
| 11 |
Hb_131466_010 |
0.0925308079 |
- |
- |
PREDICTED: putative amidase C869.01 [Jatropha curcas] |
| 12 |
Hb_000590_110 |
0.0930367944 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_006132_080 |
0.0941746489 |
- |
- |
PREDICTED: cytochrome P450 86A8-like [Jatropha curcas] |
| 14 |
Hb_002042_210 |
0.0956950586 |
- |
- |
PREDICTED: UPF0603 protein At1g54780, chloroplastic [Jatropha curcas] |
| 15 |
Hb_033187_010 |
0.0960710022 |
- |
- |
epoxide hydrolase, putative [Ricinus communis] |
| 16 |
Hb_001894_100 |
0.097561425 |
- |
- |
PREDICTED: kininogen-1-like [Jatropha curcas] |
| 17 |
Hb_041139_010 |
0.0984721552 |
- |
- |
PREDICTED: probable terpene synthase 13 [Jatropha curcas] |
| 18 |
Hb_004607_110 |
0.0998974888 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek6 [Jatropha curcas] |
| 19 |
Hb_003151_020 |
0.1006009381 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 20 |
Hb_002805_280 |
0.1021833501 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |