| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
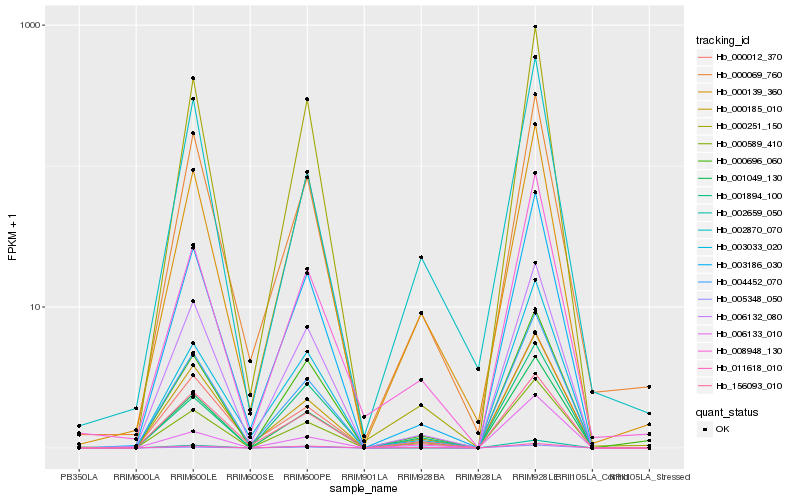
Hb_000589_410 |
0.0 |
- |
- |
PREDICTED: thylakoid lumenal 15 kDa protein 1, chloroplastic-like isoform X1 [Populus euphratica] |
| 2 |
Hb_002659_050 |
0.056943592 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 10 isoform X3 [Jatropha curcas] |
| 3 |
Hb_000185_010 |
0.084025001 |
- |
- |
Alliin lyase precursor, putative [Ricinus communis] |
| 4 |
Hb_011618_010 |
0.0882272534 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g59700 [Jatropha curcas] |
| 5 |
Hb_006132_080 |
0.09030097 |
- |
- |
PREDICTED: cytochrome P450 86A8-like [Jatropha curcas] |
| 6 |
Hb_003186_030 |
0.0911569046 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0012s05720g [Populus trichocarpa] |
| 7 |
Hb_005348_050 |
0.0919448547 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 8 |
Hb_000696_060 |
0.0973981981 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000069_760 |
0.0986739854 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1A, chloroplastic [Jatropha curcas] |
| 10 |
Hb_156093_010 |
0.1044851022 |
- |
- |
hypothetical protein RCOM_0504250 [Ricinus communis] |
| 11 |
Hb_008948_130 |
0.1047682335 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002870_070 |
0.1051436526 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 13 |
Hb_000012_370 |
0.1054950801 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000139_360 |
0.1099357024 |
- |
- |
PREDICTED: uncharacterized protein LOC105631933 [Jatropha curcas] |
| 15 |
Hb_003033_020 |
0.1120603205 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9-like [Jatropha curcas] |
| 16 |
Hb_001894_100 |
0.1124451991 |
- |
- |
PREDICTED: kininogen-1-like [Jatropha curcas] |
| 17 |
Hb_000251_150 |
0.1133872149 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 18 |
Hb_006133_010 |
0.1147721002 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 19 |
Hb_004452_070 |
0.1160191499 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 20 |
Hb_001049_130 |
0.1160521434 |
- |
- |
50S ribosomal protein L18, chloroplast precursor, putative [Ricinus communis] |