| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
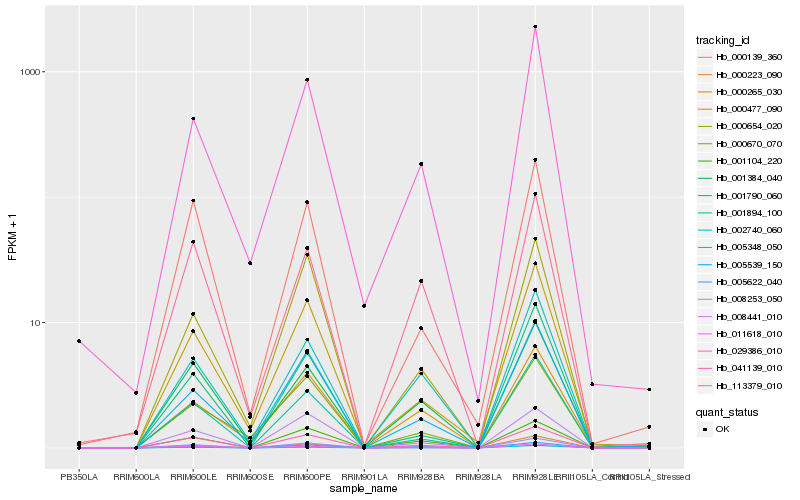
Hb_008253_050 |
0.0 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0001s36220g [Populus trichocarpa] |
| 2 |
Hb_041139_010 |
0.0546574805 |
- |
- |
PREDICTED: probable terpene synthase 13 [Jatropha curcas] |
| 3 |
Hb_011618_010 |
0.069223414 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g59700 [Jatropha curcas] |
| 4 |
Hb_001894_100 |
0.0742711772 |
- |
- |
PREDICTED: kininogen-1-like [Jatropha curcas] |
| 5 |
Hb_005348_050 |
0.0861099463 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 6 |
Hb_000223_090 |
0.0892964442 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 7 |
Hb_005539_150 |
0.0905192481 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 33 [Jatropha curcas] |
| 8 |
Hb_001104_220 |
0.0936420043 |
- |
- |
PREDICTED: uncharacterized protein LOC105629453 [Jatropha curcas] |
| 9 |
Hb_005622_040 |
0.0976862165 |
- |
- |
PREDICTED: inactive rhomboid protein 1 [Jatropha curcas] |
| 10 |
Hb_000477_090 |
0.099548855 |
- |
- |
PREDICTED: uncharacterized protein LOC105639302 [Jatropha curcas] |
| 11 |
Hb_008441_010 |
0.0995726446 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 12 |
Hb_001384_040 |
0.0998767971 |
- |
- |
PREDICTED: beta-glucosidase 17-like [Jatropha curcas] |
| 13 |
Hb_002740_060 |
0.103787029 |
- |
- |
PREDICTED: pectin acetylesterase 12 [Jatropha curcas] |
| 14 |
Hb_029386_010 |
0.1150861426 |
- |
- |
RALF-LIKE 27 family protein [Populus trichocarpa] |
| 15 |
Hb_000654_020 |
0.1178803658 |
- |
- |
hypothetical protein CICLE_v10006262mg [Citrus clementina] |
| 16 |
Hb_001790_060 |
0.1184394244 |
- |
- |
PREDICTED: uncharacterized protein LOC105644961 [Jatropha curcas] |
| 17 |
Hb_000139_360 |
0.1186766044 |
- |
- |
PREDICTED: uncharacterized protein LOC105631933 [Jatropha curcas] |
| 18 |
Hb_000265_030 |
0.1198849856 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 19 |
Hb_113379_010 |
0.1208056602 |
- |
- |
PREDICTED: beta-xylosidase/alpha-L-arabinofuranosidase 1 [Jatropha curcas] |
| 20 |
Hb_000670_070 |
0.1257351357 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |