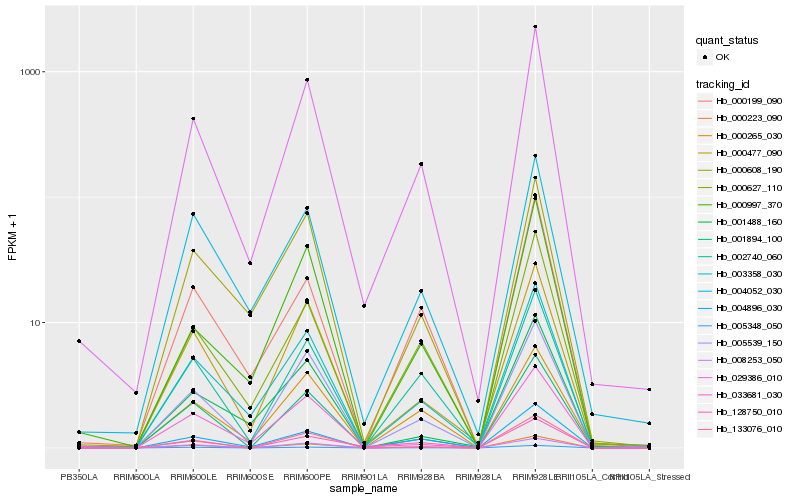
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029386_010 |
0.0 |
- |
- |
RALF-LIKE 27 family protein [Populus trichocarpa] |
| 2 |
Hb_003358_030 |
0.0727565338 |
- |
- |
ankyrin-kinase, putative [Ricinus communis] |
| 3 |
Hb_005539_150 |
0.0887839222 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 33 [Jatropha curcas] |
| 4 |
Hb_000477_090 |
0.0899796132 |
- |
- |
PREDICTED: uncharacterized protein LOC105639302 [Jatropha curcas] |
| 5 |
Hb_000627_110 |
0.0915882076 |
- |
- |
cellulose synthase, putative [Ricinus communis] |
| 6 |
Hb_000223_090 |
0.0935185683 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 7 |
Hb_004896_030 |
0.0988467988 |
- |
- |
hypothetical protein VITISV_007475 [Vitis vinifera] |
| 8 |
Hb_002740_060 |
0.1014742706 |
- |
- |
PREDICTED: pectin acetylesterase 12 [Jatropha curcas] |
| 9 |
Hb_000997_370 |
0.1108542709 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Jatropha curcas] |
| 10 |
Hb_005348_050 |
0.11146188 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 11 |
Hb_008253_050 |
0.1150861426 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0001s36220g [Populus trichocarpa] |
| 12 |
Hb_004052_030 |
0.1169893563 |
- |
- |
PREDICTED: uncharacterized protein LOC105640637 [Jatropha curcas] |
| 13 |
Hb_033681_030 |
0.1187539476 |
desease resistance |
Gene Name: NB-ARC |
Leucine-rich repeat containing protein [Theobroma cacao] |
| 14 |
Hb_000265_030 |
0.1188370902 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 15 |
Hb_000608_190 |
0.1197695148 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
PREDICTED: isopentenyl-diphosphate Delta-isomerase I [Jatropha curcas] |
| 16 |
Hb_000199_090 |
0.1199991944 |
- |
- |
Tetrapyrrole-binding family protein [Populus trichocarpa] |
| 17 |
Hb_001894_100 |
0.1208440093 |
- |
- |
PREDICTED: kininogen-1-like [Jatropha curcas] |
| 18 |
Hb_128750_010 |
0.1224300046 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 19 |
Hb_133076_010 |
0.1260726526 |
- |
- |
PREDICTED: scopoletin glucosyltransferase-like [Populus euphratica] |
| 20 |
Hb_001488_160 |
0.126977809 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |