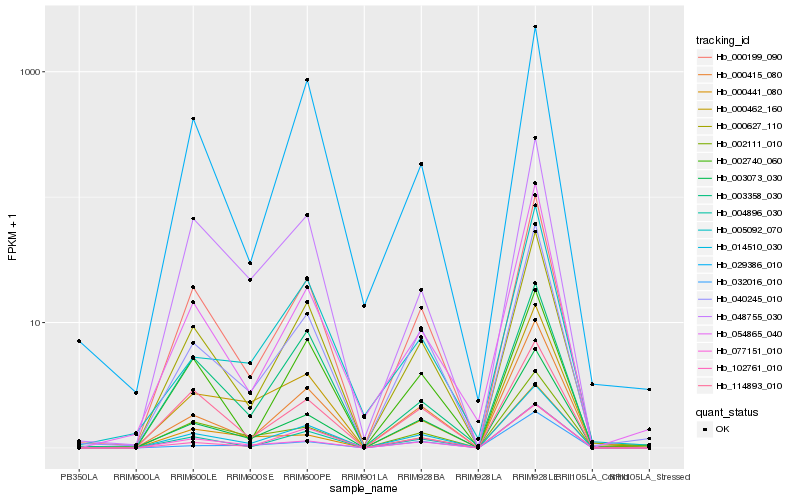
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_014510_030 |
0.0 |
- |
- |
hypothetical protein VITISV_027576 [Vitis vinifera] |
| 2 |
Hb_000199_090 |
0.0549308062 |
- |
- |
Tetrapyrrole-binding family protein [Populus trichocarpa] |
| 3 |
Hb_000627_110 |
0.055189496 |
- |
- |
cellulose synthase, putative [Ricinus communis] |
| 4 |
Hb_003073_030 |
0.0662220197 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004896_030 |
0.0679321224 |
- |
- |
hypothetical protein VITISV_007475 [Vitis vinifera] |
| 6 |
Hb_040245_010 |
0.0747605418 |
- |
- |
hypothetical protein CISIN_1g039575mg, partial [Citrus sinensis] |
| 7 |
Hb_000415_080 |
0.0802543279 |
- |
- |
- |
| 8 |
Hb_102761_010 |
0.0980517568 |
- |
- |
hypothetical protein JCGZ_21614 [Jatropha curcas] |
| 9 |
Hb_002111_010 |
0.1031406282 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 10 |
Hb_077151_010 |
0.1072370938 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 11 |
Hb_114893_010 |
0.1130225798 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 12 |
Hb_048755_030 |
0.1146922324 |
- |
- |
Periplasmic beta-glucosidase precursor, putative [Ricinus communis] |
| 13 |
Hb_003358_030 |
0.1220547595 |
- |
- |
ankyrin-kinase, putative [Ricinus communis] |
| 14 |
Hb_002740_060 |
0.1249845699 |
- |
- |
PREDICTED: pectin acetylesterase 12 [Jatropha curcas] |
| 15 |
Hb_029386_010 |
0.1281582198 |
- |
- |
RALF-LIKE 27 family protein [Populus trichocarpa] |
| 16 |
Hb_000462_160 |
0.1342709401 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 17 |
Hb_005092_070 |
0.1377288966 |
- |
- |
Oligopeptide transporter, putative [Ricinus communis] |
| 18 |
Hb_000441_080 |
0.137897347 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 19 |
Hb_032016_010 |
0.138158038 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At3g28040-like isoform X1 [Citrus sinensis] |
| 20 |
Hb_054865_040 |
0.1385718269 |
- |
- |
PREDICTED: auxin-induced protein 15A [Jatropha curcas] |