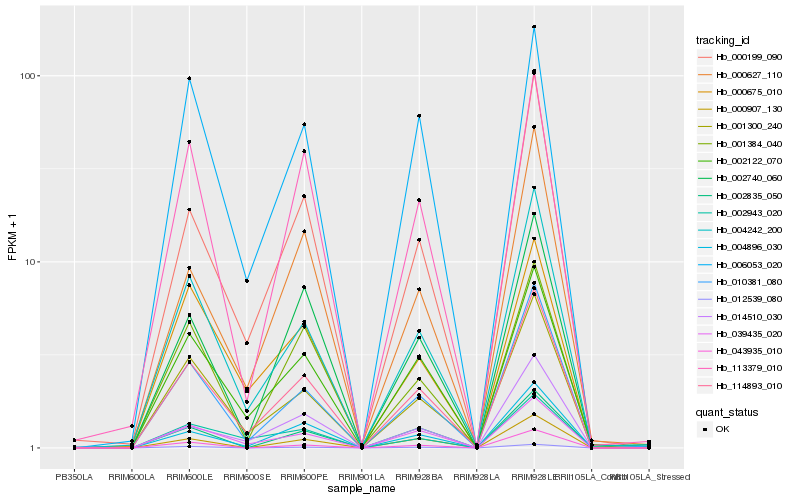
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_114893_010 |
0.0 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 2 |
Hb_001300_240 |
0.0693105979 |
- |
- |
shikimate dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_002122_070 |
0.0720981663 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_039435_020 |
0.0723712691 |
- |
- |
Lysosomal Pro-X carboxypeptidase, putative [Ricinus communis] |
| 5 |
Hb_004242_200 |
0.0739628795 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0001s44130g [Populus trichocarpa] |
| 6 |
Hb_010381_080 |
0.0779285434 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 7 |
Hb_000199_090 |
0.0915672735 |
- |
- |
Tetrapyrrole-binding family protein [Populus trichocarpa] |
| 8 |
Hb_004896_030 |
0.0962655015 |
- |
- |
hypothetical protein VITISV_007475 [Vitis vinifera] |
| 9 |
Hb_113379_010 |
0.101603802 |
- |
- |
PREDICTED: beta-xylosidase/alpha-L-arabinofuranosidase 1 [Jatropha curcas] |
| 10 |
Hb_012539_080 |
0.1087957426 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 11 |
Hb_002740_060 |
0.1098742257 |
- |
- |
PREDICTED: pectin acetylesterase 12 [Jatropha curcas] |
| 12 |
Hb_006053_020 |
0.1099231337 |
- |
- |
CAP [Theobroma cacao] |
| 13 |
Hb_001384_040 |
0.1112591804 |
- |
- |
PREDICTED: beta-glucosidase 17-like [Jatropha curcas] |
| 14 |
Hb_000627_110 |
0.1118823381 |
- |
- |
cellulose synthase, putative [Ricinus communis] |
| 15 |
Hb_000675_010 |
0.1124965329 |
- |
- |
PREDICTED: cytokinin dehydrogenase 7 [Jatropha curcas] |
| 16 |
Hb_014510_030 |
0.1130225798 |
- |
- |
hypothetical protein VITISV_027576 [Vitis vinifera] |
| 17 |
Hb_000907_130 |
0.1139294153 |
- |
- |
- |
| 18 |
Hb_002835_050 |
0.1143393099 |
- |
- |
PREDICTED: high-affinity nitrate transporter 3.1-like [Jatropha curcas] |
| 19 |
Hb_043935_010 |
0.1165160501 |
- |
- |
Receptor protein kinase-like protein [Medicago truncatula] |
| 20 |
Hb_002943_020 |
0.1172242345 |
- |
- |
hypothetical protein POPTR_0075s00240g, partial [Populus trichocarpa] |