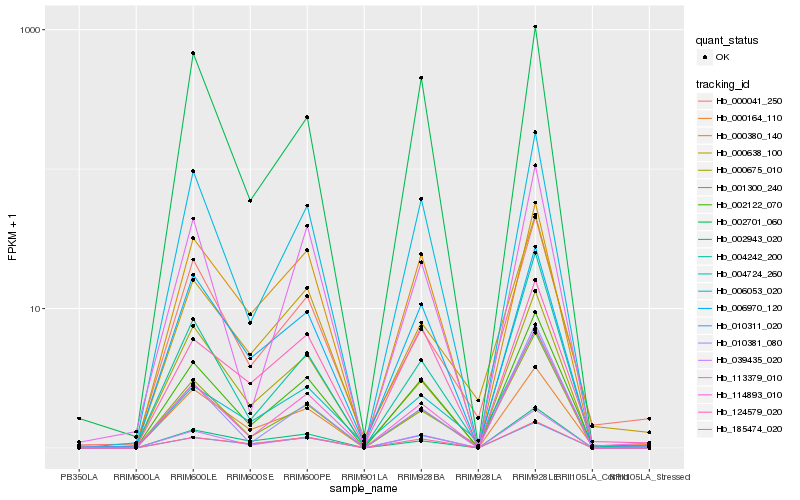
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002122_070 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_039435_020 |
0.050800078 |
- |
- |
Lysosomal Pro-X carboxypeptidase, putative [Ricinus communis] |
| 3 |
Hb_114893_010 |
0.0720981663 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 4 |
Hb_006053_020 |
0.0816998478 |
- |
- |
CAP [Theobroma cacao] |
| 5 |
Hb_185474_020 |
0.0963280274 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 6 |
Hb_001300_240 |
0.1004804462 |
- |
- |
shikimate dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_000675_010 |
0.1062050541 |
- |
- |
PREDICTED: cytokinin dehydrogenase 7 [Jatropha curcas] |
| 8 |
Hb_004242_200 |
0.1091354841 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0001s44130g [Populus trichocarpa] |
| 9 |
Hb_000164_110 |
0.1092926808 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0001s44130g [Populus trichocarpa] |
| 10 |
Hb_006970_120 |
0.1105928716 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004724_260 |
0.1118219244 |
- |
- |
PREDICTED: 40S ribosomal protein S14-2-like [Gossypium raimondii] |
| 12 |
Hb_010311_020 |
0.1162155174 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 13 |
Hb_124579_020 |
0.1173302944 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 14 |
Hb_113379_010 |
0.118888297 |
- |
- |
PREDICTED: beta-xylosidase/alpha-L-arabinofuranosidase 1 [Jatropha curcas] |
| 15 |
Hb_002943_020 |
0.1204375566 |
- |
- |
hypothetical protein POPTR_0075s00240g, partial [Populus trichocarpa] |
| 16 |
Hb_000638_100 |
0.1208638283 |
- |
- |
PREDICTED: histone deacetylase 14 [Jatropha curcas] |
| 17 |
Hb_000380_140 |
0.122503101 |
- |
- |
PREDICTED: uncharacterized protein LOC105628069 [Jatropha curcas] |
| 18 |
Hb_010381_080 |
0.1233098903 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 19 |
Hb_002701_060 |
0.12825405 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000041_250 |
0.1289190723 |
- |
- |
PREDICTED: uncharacterized protein LOC105641253 [Jatropha curcas] |