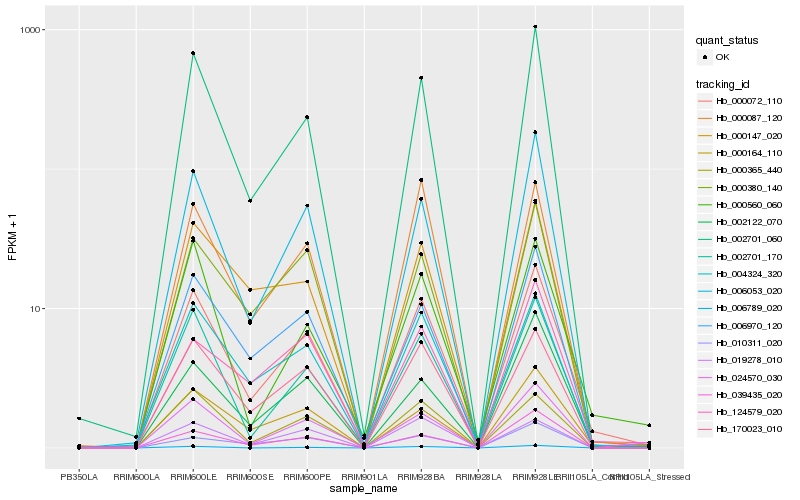
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000072_110 |
0.0 |
- |
- |
PREDICTED: U-box domain-containing protein 35 [Jatropha curcas] |
| 2 |
Hb_002701_060 |
0.0848655768 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002701_170 |
0.0952785481 |
- |
- |
PREDICTED: cytochrome P450 85A [Jatropha curcas] |
| 4 |
Hb_004324_320 |
0.0999676654 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |
| 5 |
Hb_170023_010 |
0.1053471263 |
- |
- |
hypothetical protein EUGRSUZ_C03837 [Eucalyptus grandis] |
| 6 |
Hb_006970_120 |
0.1077613097 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_006053_020 |
0.1083959291 |
- |
- |
CAP [Theobroma cacao] |
| 8 |
Hb_000164_110 |
0.1239694839 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0001s44130g [Populus trichocarpa] |
| 9 |
Hb_000365_440 |
0.1242078568 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_000087_120 |
0.1250154261 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 11 |
Hb_000147_020 |
0.1263284716 |
- |
- |
PREDICTED: uncharacterized protein LOC105638303 [Jatropha curcas] |
| 12 |
Hb_024570_030 |
0.1265502877 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 13 |
Hb_000560_060 |
0.1269017878 |
- |
- |
PREDICTED: endochitinase PR4-like [Jatropha curcas] |
| 14 |
Hb_000380_140 |
0.1283509102 |
- |
- |
PREDICTED: uncharacterized protein LOC105628069 [Jatropha curcas] |
| 15 |
Hb_006789_020 |
0.1379264447 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: wall-associated receptor kinase-like 22 [Camelina sativa] |
| 16 |
Hb_002122_070 |
0.1384834549 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_039435_020 |
0.1389286979 |
- |
- |
Lysosomal Pro-X carboxypeptidase, putative [Ricinus communis] |
| 18 |
Hb_010311_020 |
0.1420486932 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 19 |
Hb_124579_020 |
0.1431355575 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 20 |
Hb_019278_010 |
0.1435165776 |
- |
- |
PREDICTED: uncharacterized protein LOC105638282 [Jatropha curcas] |