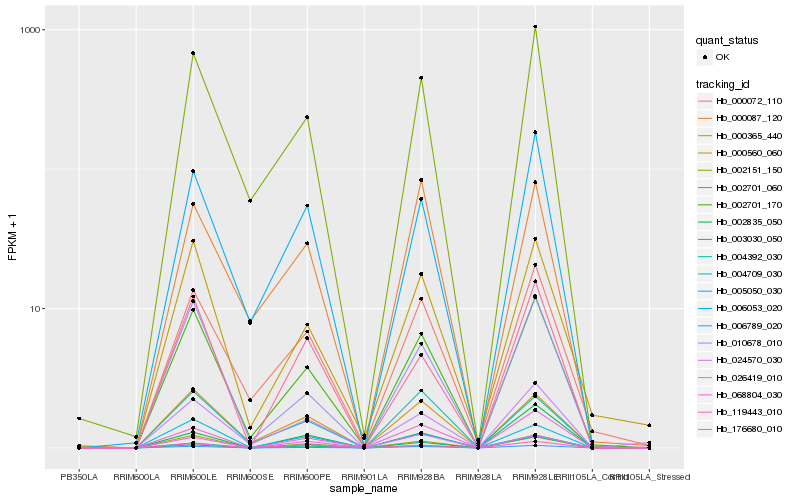
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006789_020 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: wall-associated receptor kinase-like 22 [Camelina sativa] |
| 2 |
Hb_002701_170 |
0.0913860382 |
- |
- |
PREDICTED: cytochrome P450 85A [Jatropha curcas] |
| 3 |
Hb_068804_030 |
0.0918244272 |
- |
- |
PREDICTED: uncharacterized protein LOC105639932 [Jatropha curcas] |
| 4 |
Hb_002151_150 |
0.1218906868 |
- |
- |
PREDICTED: uncharacterized protein LOC102707106 [Oryza brachyantha] |
| 5 |
Hb_002701_060 |
0.122489477 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_010678_010 |
0.1327174143 |
- |
- |
PREDICTED: alpha-glucosidase-like [Jatropha curcas] |
| 7 |
Hb_026419_010 |
0.1366362082 |
- |
- |
PREDICTED: cyclin-D3-1-like [Jatropha curcas] |
| 8 |
Hb_000072_110 |
0.1379264447 |
- |
- |
PREDICTED: U-box domain-containing protein 35 [Jatropha curcas] |
| 9 |
Hb_000560_060 |
0.1394829639 |
- |
- |
PREDICTED: endochitinase PR4-like [Jatropha curcas] |
| 10 |
Hb_119443_010 |
0.1402969731 |
- |
- |
hypothetical protein JCGZ_17348 [Jatropha curcas] |
| 11 |
Hb_006053_020 |
0.1432739606 |
- |
- |
CAP [Theobroma cacao] |
| 12 |
Hb_000365_440 |
0.1566235131 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 13 |
Hb_024570_030 |
0.1591885754 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 14 |
Hb_004709_030 |
0.1605561629 |
- |
- |
PREDICTED: receptor-like protein 12 [Vitis vinifera] |
| 15 |
Hb_004392_030 |
0.1609517794 |
- |
- |
oxidoreductase family protein [Populus trichocarpa] |
| 16 |
Hb_002835_050 |
0.1614219908 |
- |
- |
PREDICTED: high-affinity nitrate transporter 3.1-like [Jatropha curcas] |
| 17 |
Hb_005050_030 |
0.1618951663 |
- |
- |
PREDICTED: wall-associated receptor kinase 2-like [Jatropha curcas] |
| 18 |
Hb_000087_120 |
0.1646041833 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 19 |
Hb_003030_050 |
0.1648876495 |
- |
- |
- |
| 20 |
Hb_176680_010 |
0.1654757481 |
- |
- |
hypothetical protein JCGZ_22787 [Jatropha curcas] |