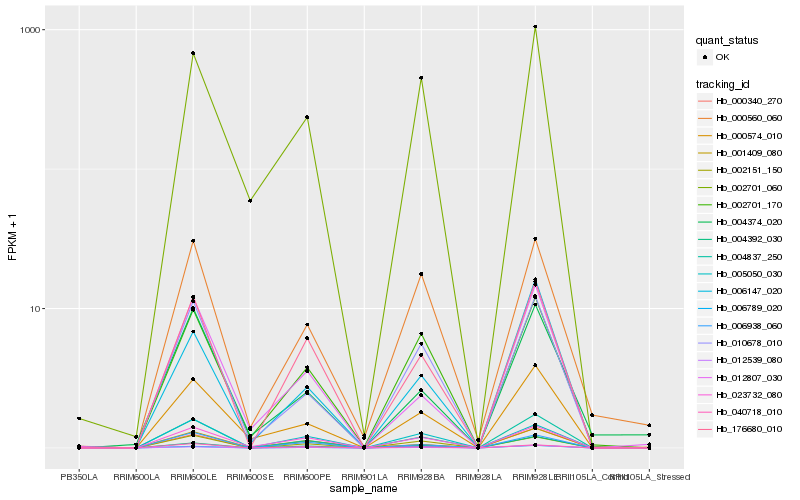
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004837_250 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_010678_010 |
0.1236154674 |
- |
- |
PREDICTED: alpha-glucosidase-like [Jatropha curcas] |
| 3 |
Hb_176680_010 |
0.1315843724 |
- |
- |
hypothetical protein JCGZ_22787 [Jatropha curcas] |
| 4 |
Hb_000574_010 |
0.1475279521 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_06096 [Jatropha curcas] |
| 5 |
Hb_023732_080 |
0.148567089 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 6 |
Hb_002701_170 |
0.1517686144 |
- |
- |
PREDICTED: cytochrome P450 85A [Jatropha curcas] |
| 7 |
Hb_006938_060 |
0.1522821158 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 8 |
Hb_004392_030 |
0.1536033821 |
- |
- |
oxidoreductase family protein [Populus trichocarpa] |
| 9 |
Hb_040718_010 |
0.1548396765 |
- |
- |
PREDICTED: beta-glucosidase 18-like [Jatropha curcas] |
| 10 |
Hb_002151_150 |
0.1550498553 |
- |
- |
PREDICTED: uncharacterized protein LOC102707106 [Oryza brachyantha] |
| 11 |
Hb_001409_080 |
0.155829174 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 isoform X1 [Populus euphratica] |
| 12 |
Hb_012807_030 |
0.1637872099 |
- |
- |
PREDICTED: uncharacterized protein LOC105628042 isoform X1 [Jatropha curcas] |
| 13 |
Hb_012539_080 |
0.1638051718 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 14 |
Hb_000340_270 |
0.1645523239 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 15 |
Hb_000560_060 |
0.1678220708 |
- |
- |
PREDICTED: endochitinase PR4-like [Jatropha curcas] |
| 16 |
Hb_002701_060 |
0.170162455 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_006789_020 |
0.1712473256 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: wall-associated receptor kinase-like 22 [Camelina sativa] |
| 18 |
Hb_006147_020 |
0.1720035628 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004374_020 |
0.1725151486 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog MMK2-like [Jatropha curcas] |
| 20 |
Hb_005050_030 |
0.1725162399 |
- |
- |
PREDICTED: wall-associated receptor kinase 2-like [Jatropha curcas] |