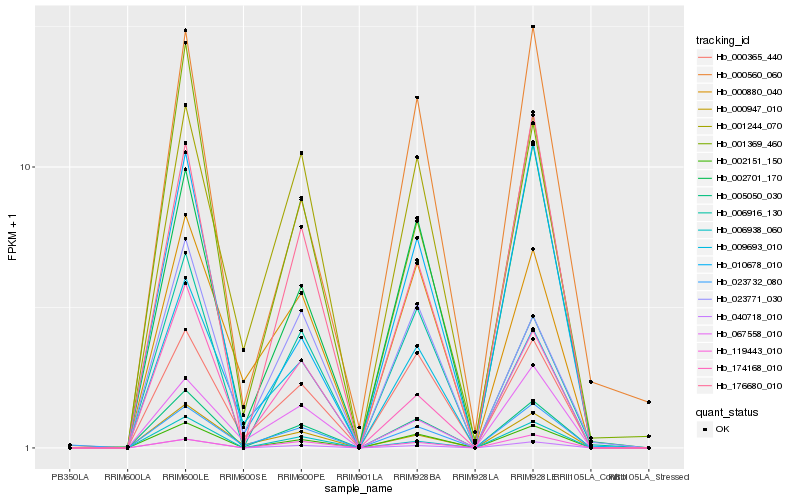
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005050_030 |
0.0 |
- |
- |
PREDICTED: wall-associated receptor kinase 2-like [Jatropha curcas] |
| 2 |
Hb_002151_150 |
0.0612322792 |
- |
- |
PREDICTED: uncharacterized protein LOC102707106 [Oryza brachyantha] |
| 3 |
Hb_040718_010 |
0.0874220365 |
- |
- |
PREDICTED: beta-glucosidase 18-like [Jatropha curcas] |
| 4 |
Hb_000365_440 |
0.0896218866 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 5 |
Hb_002701_170 |
0.1052379449 |
- |
- |
PREDICTED: cytochrome P450 85A [Jatropha curcas] |
| 6 |
Hb_000947_010 |
0.1069838667 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 7 |
Hb_009693_010 |
0.1099227377 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Gossypium raimondii] |
| 8 |
Hb_001244_070 |
0.1204765858 |
- |
- |
hypothetical protein POPTR_0004s13470g [Populus trichocarpa] |
| 9 |
Hb_006916_130 |
0.1207542354 |
- |
- |
PREDICTED: cytochrome P450 85A [Jatropha curcas] |
| 10 |
Hb_174168_010 |
0.1240736378 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 11 |
Hb_023732_080 |
0.1256686055 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 12 |
Hb_001369_460 |
0.127937746 |
- |
- |
vacuolar invertase [Manihot esculenta] |
| 13 |
Hb_006938_060 |
0.1283888926 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 14 |
Hb_000560_060 |
0.1315991136 |
- |
- |
PREDICTED: endochitinase PR4-like [Jatropha curcas] |
| 15 |
Hb_000880_040 |
0.1331534962 |
- |
- |
- |
| 16 |
Hb_176680_010 |
0.1349521154 |
- |
- |
hypothetical protein JCGZ_22787 [Jatropha curcas] |
| 17 |
Hb_010678_010 |
0.1388625626 |
- |
- |
PREDICTED: alpha-glucosidase-like [Jatropha curcas] |
| 18 |
Hb_023771_030 |
0.1396724553 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_067558_010 |
0.140896827 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 20 |
Hb_119443_010 |
0.1413696718 |
- |
- |
hypothetical protein JCGZ_17348 [Jatropha curcas] |