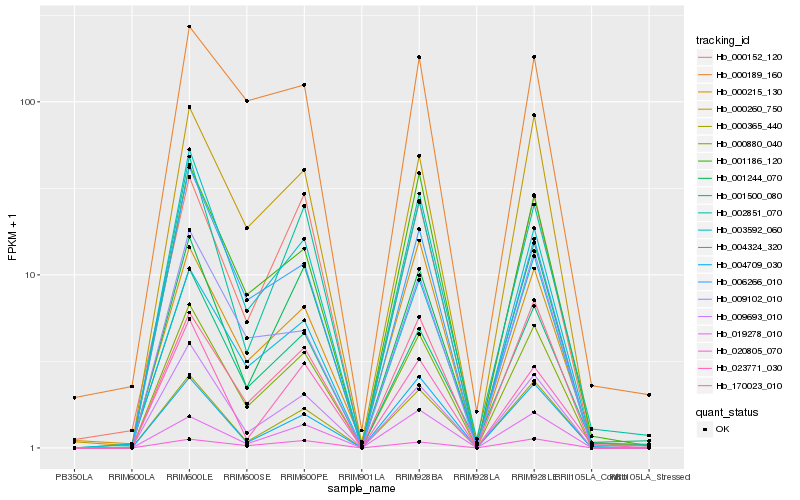
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000880_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_009693_010 |
0.067983667 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Gossypium raimondii] |
| 3 |
Hb_000260_750 |
0.073692523 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 7-like [Jatropha curcas] |
| 4 |
Hb_000365_440 |
0.079539945 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 5 |
Hb_001244_070 |
0.0826922367 |
- |
- |
hypothetical protein POPTR_0004s13470g [Populus trichocarpa] |
| 6 |
Hb_170023_010 |
0.0907268783 |
- |
- |
hypothetical protein EUGRSUZ_C03837 [Eucalyptus grandis] |
| 7 |
Hb_002851_070 |
0.098866337 |
- |
- |
PREDICTED: uncharacterized protein LOC105646410 [Jatropha curcas] |
| 8 |
Hb_000152_120 |
0.1006025282 |
- |
- |
alpha-l-fucosidase, putative [Ricinus communis] |
| 9 |
Hb_001186_120 |
0.1009740621 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase 5 [Jatropha curcas] |
| 10 |
Hb_004324_320 |
0.1036132006 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |
| 11 |
Hb_001500_080 |
0.1049006764 |
- |
- |
PREDICTED: uncharacterized protein LOC105647337 isoform X1 [Jatropha curcas] |
| 12 |
Hb_009102_010 |
0.1095702024 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_020805_070 |
0.1100348133 |
- |
- |
PREDICTED: MLO protein homolog 1-like [Jatropha curcas] |
| 14 |
Hb_003592_060 |
0.1190636843 |
- |
- |
hypothetical protein F383_11017 [Gossypium arboreum] |
| 15 |
Hb_006266_010 |
0.1209035595 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_019278_010 |
0.1235409038 |
- |
- |
PREDICTED: uncharacterized protein LOC105638282 [Jatropha curcas] |
| 17 |
Hb_004709_030 |
0.1248387857 |
- |
- |
PREDICTED: receptor-like protein 12 [Vitis vinifera] |
| 18 |
Hb_000215_130 |
0.1254817734 |
- |
- |
PREDICTED: probable pectinesterase 53 [Jatropha curcas] |
| 19 |
Hb_000189_160 |
0.1311953476 |
- |
- |
PREDICTED: GDP-mannose 3,5-epimerase 2 [Eucalyptus grandis] |
| 20 |
Hb_023771_030 |
0.1319142938 |
- |
- |
unnamed protein product [Vitis vinifera] |