| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
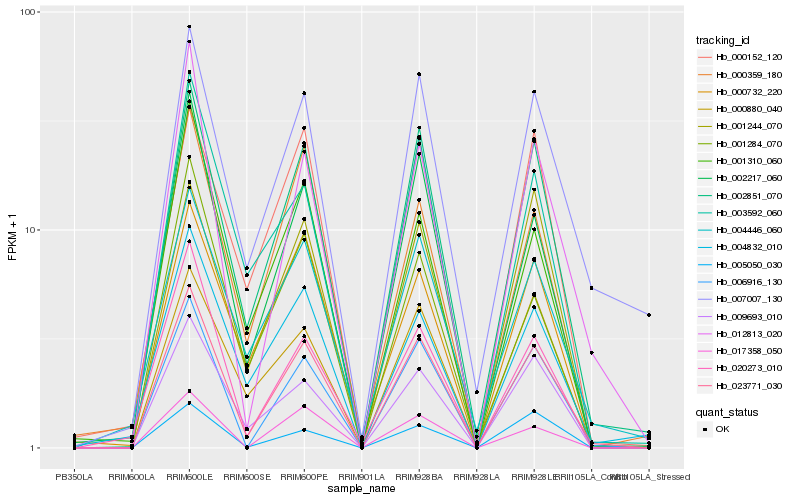
Hb_023771_030 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_002851_070 |
0.0627409249 |
- |
- |
PREDICTED: uncharacterized protein LOC105646410 [Jatropha curcas] |
| 3 |
Hb_006916_130 |
0.0827303968 |
- |
- |
PREDICTED: cytochrome P450 85A [Jatropha curcas] |
| 4 |
Hb_002217_060 |
0.0980730779 |
- |
- |
PREDICTED: polygalacturonase At1g48100 [Jatropha curcas] |
| 5 |
Hb_012813_020 |
0.1023146658 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase, basic isoform-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_009693_010 |
0.1042509376 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Gossypium raimondii] |
| 7 |
Hb_003592_060 |
0.1214573143 |
- |
- |
hypothetical protein F383_11017 [Gossypium arboreum] |
| 8 |
Hb_020273_010 |
0.1258040256 |
- |
- |
PREDICTED: beta-galactosidase 3 [Jatropha curcas] |
| 9 |
Hb_007007_130 |
0.1264193289 |
transcription factor |
TF Family: MIKC |
MADS-box transcription factor 2 [Hevea brasiliensis] |
| 10 |
Hb_017358_050 |
0.128495787 |
- |
- |
PREDICTED: uncharacterized protein LOC105641552 [Jatropha curcas] |
| 11 |
Hb_004446_060 |
0.1309190259 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 12 |
Hb_000880_040 |
0.1319142938 |
- |
- |
- |
| 13 |
Hb_001310_060 |
0.1322381935 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 14 |
Hb_000732_220 |
0.1340328239 |
- |
- |
PREDICTED: importin subunit alpha-5 [Jatropha curcas] |
| 15 |
Hb_004832_010 |
0.1355867793 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000359_180 |
0.1371998364 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000152_120 |
0.1384244632 |
- |
- |
alpha-l-fucosidase, putative [Ricinus communis] |
| 18 |
Hb_001244_070 |
0.1386928515 |
- |
- |
hypothetical protein POPTR_0004s13470g [Populus trichocarpa] |
| 19 |
Hb_001284_070 |
0.1396300023 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_005050_030 |
0.1396724553 |
- |
- |
PREDICTED: wall-associated receptor kinase 2-like [Jatropha curcas] |