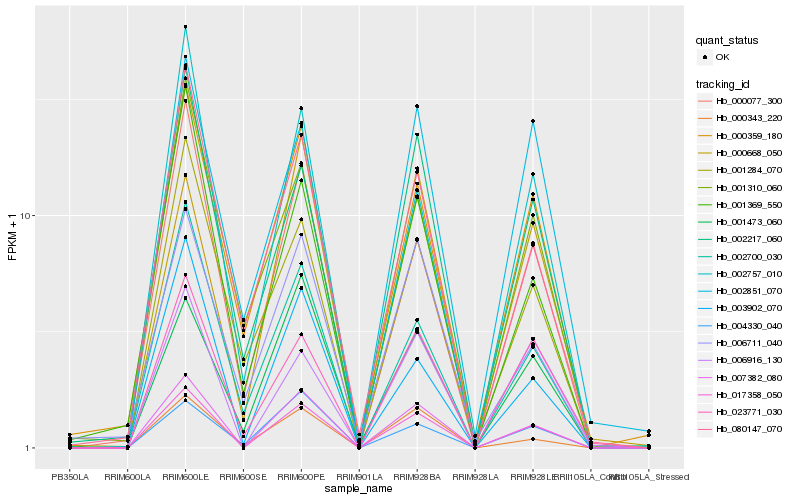
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017358_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641552 [Jatropha curcas] |
| 2 |
Hb_007382_080 |
0.0382919546 |
- |
- |
glutaredoxin, grx, putative [Ricinus communis] |
| 3 |
Hb_023771_030 |
0.128495787 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_006916_130 |
0.1320795047 |
- |
- |
PREDICTED: cytochrome P450 85A [Jatropha curcas] |
| 5 |
Hb_000359_180 |
0.1378371598 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000077_300 |
0.1402175345 |
- |
- |
PREDICTED: expansin-B3-like [Jatropha curcas] |
| 7 |
Hb_002217_060 |
0.1417971076 |
- |
- |
PREDICTED: polygalacturonase At1g48100 [Jatropha curcas] |
| 8 |
Hb_000343_220 |
0.1485355208 |
- |
- |
hypothetical protein CISIN_1g037489mg [Citrus sinensis] |
| 9 |
Hb_001473_060 |
0.152744091 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 8 [Jatropha curcas] |
| 10 |
Hb_003902_070 |
0.1531312243 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004330_040 |
0.1546045323 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002851_070 |
0.1554075749 |
- |
- |
PREDICTED: uncharacterized protein LOC105646410 [Jatropha curcas] |
| 13 |
Hb_080147_070 |
0.1568696178 |
- |
- |
Rop5 [Hevea brasiliensis] |
| 14 |
Hb_002757_010 |
0.1581825475 |
- |
- |
hypothetical protein POPTR_0013s10580g [Populus trichocarpa] |
| 15 |
Hb_002700_030 |
0.1587413992 |
- |
- |
PREDICTED: serine carboxypeptidase-like 45 [Jatropha curcas] |
| 16 |
Hb_001284_070 |
0.158750854 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001369_550 |
0.1618162298 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 18 |
Hb_006711_040 |
0.1624790779 |
- |
- |
PREDICTED: probable protein ABIL5 [Jatropha curcas] |
| 19 |
Hb_000668_050 |
0.1647126331 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 20 |
Hb_001310_060 |
0.1654493627 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |