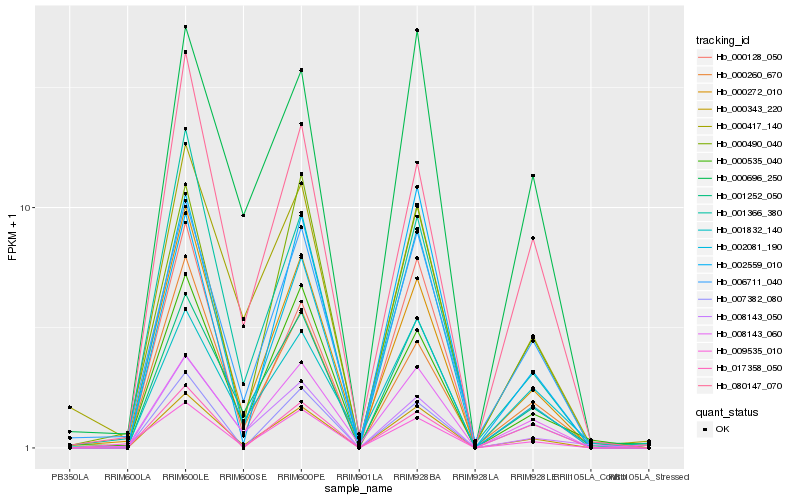
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000343_220 |
0.0 |
- |
- |
hypothetical protein CISIN_1g037489mg [Citrus sinensis] |
| 2 |
Hb_001252_050 |
0.0835976578 |
- |
- |
Wall-associated receptor kinase-like 20 [Morus notabilis] |
| 3 |
Hb_006711_040 |
0.0912370326 |
- |
- |
PREDICTED: probable protein ABIL5 [Jatropha curcas] |
| 4 |
Hb_002081_190 |
0.0947268266 |
- |
- |
hypothetical protein EUGRSUZ_I010391, partial [Eucalyptus grandis] |
| 5 |
Hb_000490_040 |
0.1049804388 |
- |
- |
PREDICTED: cyclin-D3-1 [Jatropha curcas] |
| 6 |
Hb_000272_010 |
0.1050352389 |
- |
- |
PREDICTED: DNA replication licensing factor MCM5 [Jatropha curcas] |
| 7 |
Hb_000128_050 |
0.1106425305 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_008143_060 |
0.1251864833 |
- |
- |
PREDICTED: putative hydrolase C777.06c isoform X2 [Jatropha curcas] |
| 9 |
Hb_007382_080 |
0.1258234925 |
- |
- |
glutaredoxin, grx, putative [Ricinus communis] |
| 10 |
Hb_000696_250 |
0.1259675007 |
- |
- |
PREDICTED: patellin-6 [Jatropha curcas] |
| 11 |
Hb_009535_010 |
0.1294809556 |
- |
- |
PREDICTED: DNA replication licensing factor MCM4 [Jatropha curcas] |
| 12 |
Hb_000535_040 |
0.1298911231 |
- |
- |
PREDICTED: uncharacterized protein LOC105642104 [Jatropha curcas] |
| 13 |
Hb_002559_010 |
0.1303181553 |
- |
- |
hypothetical protein JCGZ_06274 [Jatropha curcas] |
| 14 |
Hb_001366_380 |
0.1362679236 |
- |
- |
PREDICTED: pectin acetylesterase 6-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_008143_050 |
0.1389703207 |
- |
- |
PREDICTED: cytochrome P450 710A1-like [Jatropha curcas] |
| 16 |
Hb_001832_140 |
0.1391061027 |
- |
- |
endomembrane protein 70 family protein [Medicago truncatula] |
| 17 |
Hb_080147_070 |
0.1411135285 |
- |
- |
Rop5 [Hevea brasiliensis] |
| 18 |
Hb_000260_670 |
0.1415396777 |
- |
- |
PREDICTED: serine/threonine-protein kinase TIO isoform X1 [Jatropha curcas] |
| 19 |
Hb_000417_140 |
0.1452344413 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 2 [Jatropha curcas] |
| 20 |
Hb_017358_050 |
0.1485355208 |
- |
- |
PREDICTED: uncharacterized protein LOC105641552 [Jatropha curcas] |