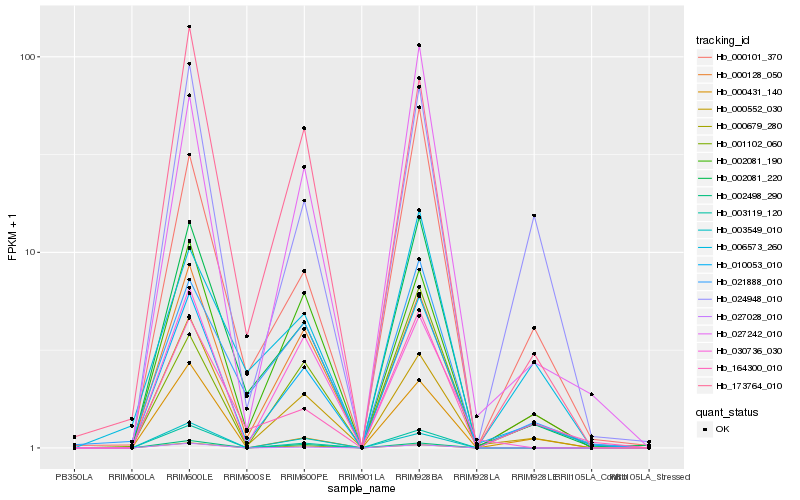
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010053_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0515s00220g [Populus trichocarpa] |
| 2 |
Hb_000128_050 |
0.1108176593 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002081_220 |
0.1130609166 |
- |
- |
PREDICTED: uncharacterized protein LOC103707552 [Phoenix dactylifera] |
| 4 |
Hb_000552_030 |
0.1192598192 |
- |
- |
glucose-6-phosphate 1-dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_024948_010 |
0.125839668 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase, basic isoform-like [Populus euphratica] |
| 6 |
Hb_164300_010 |
0.1346991791 |
- |
- |
PREDICTED: thaumatin-like protein 1b [Jatropha curcas] |
| 7 |
Hb_002081_190 |
0.1350116861 |
- |
- |
hypothetical protein EUGRSUZ_I010391, partial [Eucalyptus grandis] |
| 8 |
Hb_027242_010 |
0.1423965762 |
- |
- |
PREDICTED: pathogenesis-related protein 1-like [Populus euphratica] |
| 9 |
Hb_173764_010 |
0.1474067273 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 10 |
Hb_003119_120 |
0.1550024169 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Nicotiana sylvestris] |
| 11 |
Hb_000101_370 |
0.1583751871 |
- |
- |
PREDICTED: dihydrofolate reductase-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_001102_060 |
0.1595076653 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase At1g78530 isoform X1 [Jatropha curcas] |
| 13 |
Hb_021888_010 |
0.1609171284 |
- |
- |
PREDICTED: aspartic proteinase-like protein 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_027028_010 |
0.1653114208 |
- |
- |
hypothetical protein JCGZ_01141 [Jatropha curcas] |
| 15 |
Hb_000431_140 |
0.1724532105 |
- |
- |
PREDICTED: acetolactate synthase 1, chloroplastic-like [Vitis vinifera] |
| 16 |
Hb_000679_280 |
0.1733338803 |
- |
- |
hypothetical protein POPTR_0006s27120g [Populus trichocarpa] |
| 17 |
Hb_030736_030 |
0.1747365105 |
- |
- |
hypothetical protein VITISV_006651 [Vitis vinifera] |
| 18 |
Hb_002498_290 |
0.1825577793 |
- |
- |
PREDICTED: autophagy-related protein 8C-like isoform X2 [Elaeis guineensis] |
| 19 |
Hb_003549_010 |
0.1845778256 |
- |
- |
PREDICTED: uncharacterized protein LOC105647434 [Jatropha curcas] |
| 20 |
Hb_006573_260 |
0.187400041 |
- |
- |
- |