| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
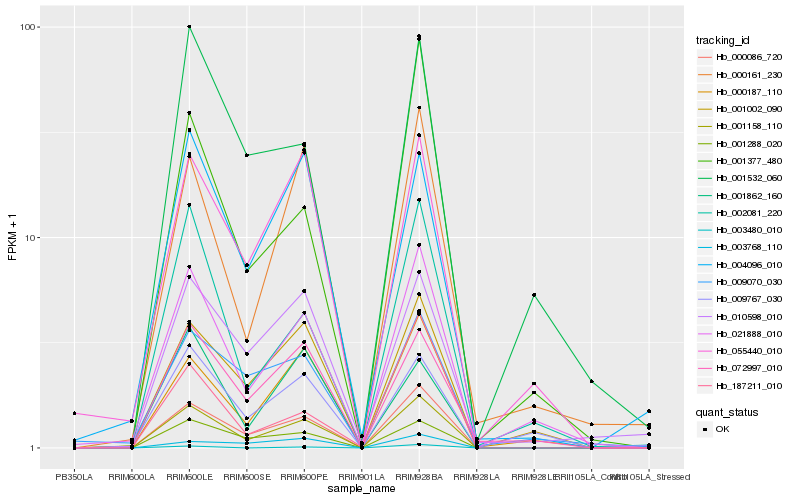
Hb_001158_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648901 [Jatropha curcas] |
| 2 |
Hb_000086_720 |
0.0803493462 |
- |
- |
leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 3 |
Hb_001288_020 |
0.1099854449 |
- |
- |
- |
| 4 |
Hb_021888_010 |
0.1203660253 |
- |
- |
PREDICTED: aspartic proteinase-like protein 1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000187_110 |
0.1331768832 |
- |
- |
PREDICTED: serine carboxypeptidase-like 7 [Jatropha curcas] |
| 6 |
Hb_001002_090 |
0.1373893065 |
- |
- |
PREDICTED: receptor protein kinase-like protein At4g34220 [Jatropha curcas] |
| 7 |
Hb_072997_010 |
0.1425828236 |
- |
- |
hypothetical protein JCGZ_18743 [Jatropha curcas] |
| 8 |
Hb_000161_230 |
0.1451284552 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 9 |
Hb_002081_220 |
0.1510708038 |
- |
- |
PREDICTED: uncharacterized protein LOC103707552 [Phoenix dactylifera] |
| 10 |
Hb_055440_010 |
0.1513077072 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 11 |
Hb_004096_010 |
0.1571668599 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 40 [Jatropha curcas] |
| 12 |
Hb_010598_010 |
0.1628855879 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |
| 13 |
Hb_009767_030 |
0.1678005055 |
- |
- |
PREDICTED: uncharacterized protein LOC105631977 [Jatropha curcas] |
| 14 |
Hb_001862_160 |
0.172762493 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.1 [Jatropha curcas] |
| 15 |
Hb_001532_060 |
0.1770574937 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 16 |
Hb_009070_030 |
0.1789593251 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 10 [Jatropha curcas] |
| 17 |
Hb_001377_480 |
0.1821451352 |
transcription factor |
TF Family: MYB |
Myb domain protein 7, putative [Theobroma cacao] |
| 18 |
Hb_003768_110 |
0.1831846123 |
- |
- |
hypothetical protein POPTR_0004s12230g [Populus trichocarpa] |
| 19 |
Hb_003480_010 |
0.1832215895 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 20 |
Hb_187211_010 |
0.1840740421 |
- |
- |
PREDICTED: beta-glucosidase 17-like [Jatropha curcas] |