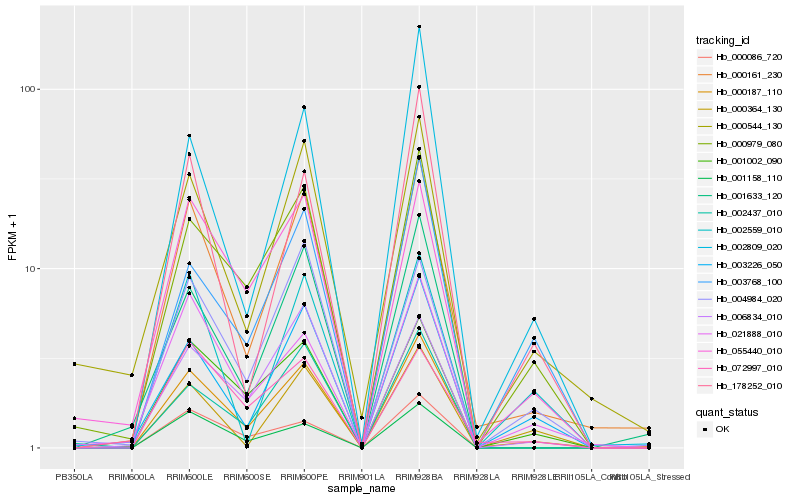
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000187_110 |
0.0 |
- |
- |
PREDICTED: serine carboxypeptidase-like 7 [Jatropha curcas] |
| 2 |
Hb_000161_230 |
0.0653392573 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 3 |
Hb_000979_080 |
0.1015625118 |
- |
- |
PREDICTED: 21 kDa protein-like [Jatropha curcas] |
| 4 |
Hb_001002_090 |
0.1084350387 |
- |
- |
PREDICTED: receptor protein kinase-like protein At4g34220 [Jatropha curcas] |
| 5 |
Hb_003226_050 |
0.1213895081 |
- |
- |
Pectinesterase-3 precursor, putative [Ricinus communis] |
| 6 |
Hb_055440_010 |
0.1282161733 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 7 |
Hb_001633_120 |
0.1311219625 |
- |
- |
PREDICTED: uncharacterized protein LOC105630199 [Jatropha curcas] |
| 8 |
Hb_001158_110 |
0.1331768832 |
- |
- |
PREDICTED: uncharacterized protein LOC105648901 [Jatropha curcas] |
| 9 |
Hb_000544_130 |
0.1337672456 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 10 |
Hb_021888_010 |
0.1360235202 |
- |
- |
PREDICTED: aspartic proteinase-like protein 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000086_720 |
0.1400471495 |
- |
- |
leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 12 |
Hb_002437_010 |
0.1425736143 |
- |
- |
PREDICTED: uncharacterized protein LOC105123193 [Populus euphratica] |
| 13 |
Hb_006834_010 |
0.1472556254 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003768_100 |
0.1478685231 |
- |
- |
hypothetical protein JCGZ_19930 [Jatropha curcas] |
| 15 |
Hb_004984_020 |
0.1478898799 |
- |
- |
PREDICTED: uncharacterized protein LOC105647814 [Jatropha curcas] |
| 16 |
Hb_000364_130 |
0.1514833729 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_178252_010 |
0.1523343144 |
- |
- |
hypothetical protein POPTR_0018s01890g [Populus trichocarpa] |
| 18 |
Hb_002809_020 |
0.1546385913 |
- |
- |
PREDICTED: leucoanthocyanidin dioxygenase [Jatropha curcas] |
| 19 |
Hb_072997_010 |
0.1583736807 |
- |
- |
hypothetical protein JCGZ_18743 [Jatropha curcas] |
| 20 |
Hb_002559_010 |
0.165815956 |
- |
- |
hypothetical protein JCGZ_06274 [Jatropha curcas] |