| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
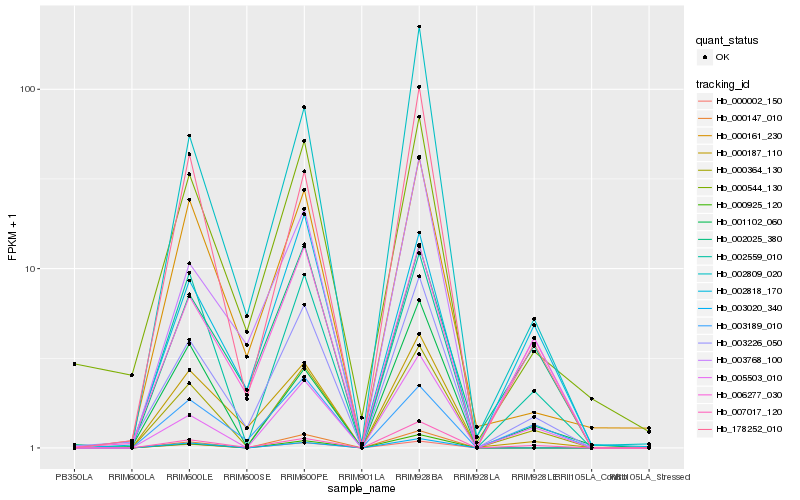
Hb_000364_130 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_002559_010 |
0.0821197792 |
- |
- |
hypothetical protein JCGZ_06274 [Jatropha curcas] |
| 3 |
Hb_003226_050 |
0.0972150716 |
- |
- |
Pectinesterase-3 precursor, putative [Ricinus communis] |
| 4 |
Hb_005503_010 |
0.1235458227 |
- |
- |
PREDICTED: probable protein Pop3 [Jatropha curcas] |
| 5 |
Hb_178252_010 |
0.143292095 |
- |
- |
hypothetical protein POPTR_0018s01890g [Populus trichocarpa] |
| 6 |
Hb_007017_120 |
0.1454632601 |
- |
- |
PREDICTED: exocyst complex component EXO84A [Jatropha curcas] |
| 7 |
Hb_006277_030 |
0.1497244371 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000187_110 |
0.1514833729 |
- |
- |
PREDICTED: serine carboxypeptidase-like 7 [Jatropha curcas] |
| 9 |
Hb_000002_150 |
0.1516211183 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001102_060 |
0.1522900686 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase At1g78530 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003768_100 |
0.1536422366 |
- |
- |
hypothetical protein JCGZ_19930 [Jatropha curcas] |
| 12 |
Hb_002025_380 |
0.1539440628 |
- |
- |
Uncharacterized protein TCM_011176 [Theobroma cacao] |
| 13 |
Hb_003020_340 |
0.1556611736 |
- |
- |
PREDICTED: uncharacterized protein LOC105631327 [Jatropha curcas] |
| 14 |
Hb_000161_230 |
0.1558753733 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 15 |
Hb_003189_010 |
0.1586250624 |
- |
- |
hypothetical protein EUGRSUZ_G02605 [Eucalyptus grandis] |
| 16 |
Hb_000544_130 |
0.1623623634 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 17 |
Hb_000147_010 |
0.1667617112 |
- |
- |
PREDICTED: UDP-glycosyltransferase 87A1-like [Jatropha curcas] |
| 18 |
Hb_002809_020 |
0.1670943655 |
- |
- |
PREDICTED: leucoanthocyanidin dioxygenase [Jatropha curcas] |
| 19 |
Hb_002818_170 |
0.1695441529 |
- |
- |
hypothetical protein POPTR_0014s10650g [Populus trichocarpa] |
| 20 |
Hb_000925_120 |
0.1704921467 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |