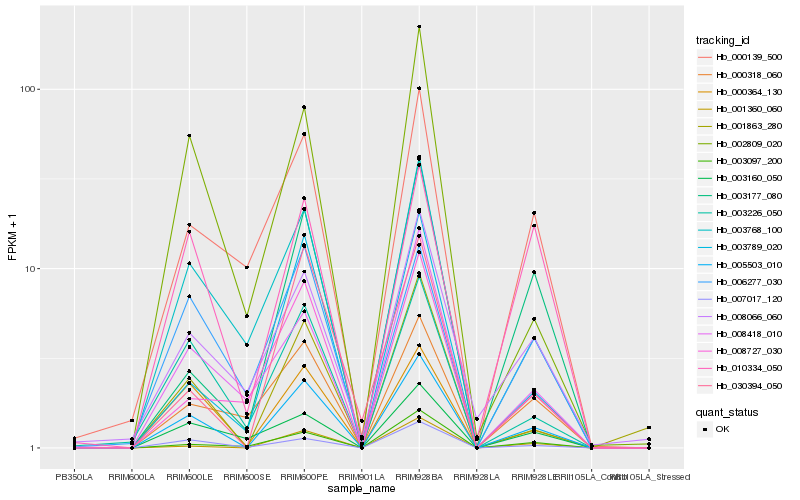
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005503_010 |
0.0 |
- |
- |
PREDICTED: probable protein Pop3 [Jatropha curcas] |
| 2 |
Hb_000364_130 |
0.1235458227 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_007017_120 |
0.1424581891 |
- |
- |
PREDICTED: exocyst complex component EXO84A [Jatropha curcas] |
| 4 |
Hb_003768_100 |
0.1459149454 |
- |
- |
hypothetical protein JCGZ_19930 [Jatropha curcas] |
| 5 |
Hb_001360_060 |
0.1485658046 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 32 [Jatropha curcas] |
| 6 |
Hb_003226_050 |
0.1487185707 |
- |
- |
Pectinesterase-3 precursor, putative [Ricinus communis] |
| 7 |
Hb_001863_280 |
0.1514227843 |
- |
- |
PREDICTED: protein EXORDIUM-like 2 [Jatropha curcas] |
| 8 |
Hb_003789_020 |
0.1699869129 |
- |
- |
hypothetical protein JCGZ_23602 [Jatropha curcas] |
| 9 |
Hb_003097_200 |
0.1711501528 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000139_500 |
0.1714802252 |
- |
- |
PREDICTED: putative uncharacterized protein DDB_G0286901 [Prunus mume] |
| 11 |
Hb_000318_060 |
0.173126926 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002809_020 |
0.1777100569 |
- |
- |
PREDICTED: leucoanthocyanidin dioxygenase [Jatropha curcas] |
| 13 |
Hb_006277_030 |
0.1785973791 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_008066_060 |
0.179839052 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 15 |
Hb_008418_010 |
0.1803067897 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH93-like [Jatropha curcas] |
| 16 |
Hb_003160_050 |
0.1803610783 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 17 |
Hb_010334_050 |
0.1805204527 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003177_080 |
0.1806362837 |
- |
- |
hypothetical protein L484_016032 [Morus notabilis] |
| 19 |
Hb_030394_050 |
0.1833780029 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 20 |
Hb_008727_030 |
0.1848848563 |
- |
- |
conserved hypothetical protein [Ricinus communis] |