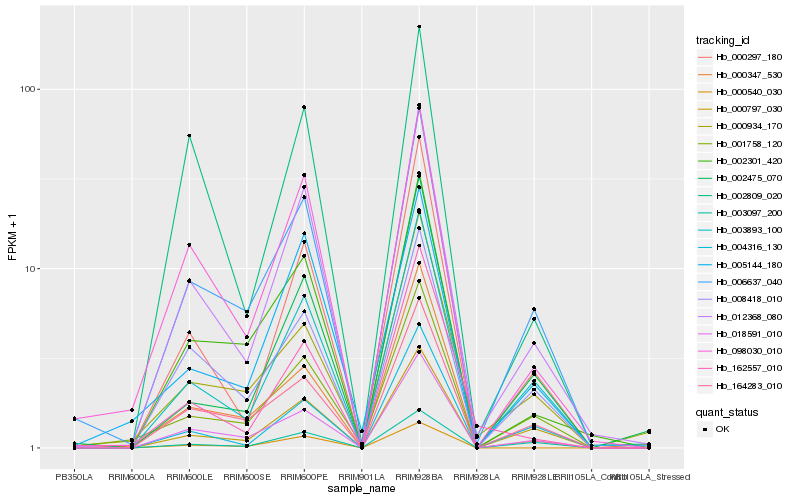
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012368_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105126277 [Populus euphratica] |
| 2 |
Hb_003893_100 |
0.0786338686 |
- |
- |
PREDICTED: uncharacterized protein LOC105648774 [Jatropha curcas] |
| 3 |
Hb_000297_180 |
0.0899960897 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |
| 4 |
Hb_006637_040 |
0.0904120369 |
- |
- |
phenylalanine ammonia-lyase 2 [Manihot esculenta] |
| 5 |
Hb_018591_010 |
0.0919287809 |
- |
- |
kinase, putative [Ricinus communis] |
| 6 |
Hb_098030_010 |
0.09575815 |
- |
- |
hypothetical protein JCGZ_06251 [Jatropha curcas] |
| 7 |
Hb_002301_420 |
0.10693677 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0011s05320g [Populus trichocarpa] |
| 8 |
Hb_008418_010 |
0.1101579218 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH93-like [Jatropha curcas] |
| 9 |
Hb_000797_030 |
0.1112793084 |
- |
- |
PREDICTED: leucoanthocyanidin dioxygenase [Jatropha curcas] |
| 10 |
Hb_164283_010 |
0.1142566623 |
- |
- |
hypothetical protein RCOM_0725500 [Ricinus communis] |
| 11 |
Hb_003097_200 |
0.1142579228 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001758_120 |
0.119442468 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_004316_130 |
0.126539952 |
- |
- |
PREDICTED: glutathione transferase GST 23-like, partial [Eucalyptus grandis] |
| 14 |
Hb_162557_010 |
0.1294333021 |
- |
- |
PREDICTED: uncharacterized protein LOC105634192 [Jatropha curcas] |
| 15 |
Hb_002809_020 |
0.1295519765 |
- |
- |
PREDICTED: leucoanthocyanidin dioxygenase [Jatropha curcas] |
| 16 |
Hb_002475_070 |
0.1307301319 |
- |
- |
hypothetical protein POPTR_0017s02440g, partial [Populus trichocarpa] |
| 17 |
Hb_000540_030 |
0.1318737082 |
- |
- |
Chitinase 1 precursor, putative [Ricinus communis] |
| 18 |
Hb_005144_180 |
0.1319497964 |
- |
- |
PREDICTED: protein enabled homolog [Populus euphratica] |
| 19 |
Hb_000347_530 |
0.1346299627 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB114-like [Jatropha curcas] |
| 20 |
Hb_000934_170 |
0.1386416767 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |