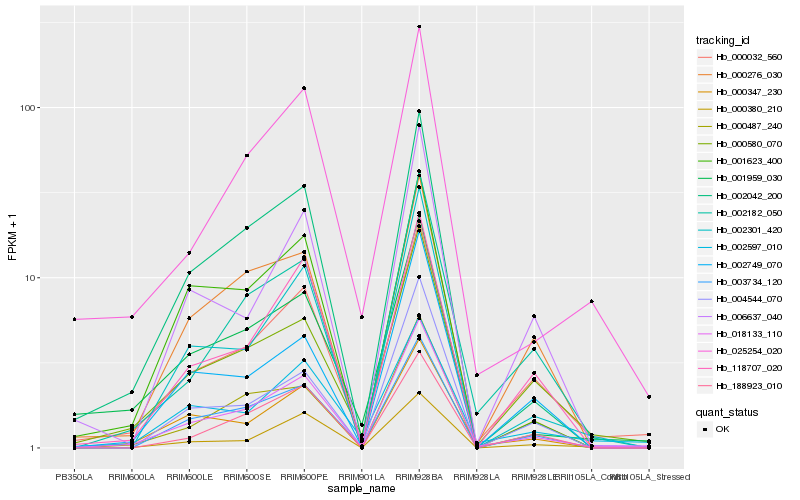
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_018133_110 |
0.0 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_000032_560 |
0.072754146 |
- |
- |
hypothetical protein JCGZ_15037 [Jatropha curcas] |
| 3 |
Hb_000347_230 |
0.0961685454 |
- |
- |
inter-alpha-trypsin inhibitor heavy chain, putative [Ricinus communis] |
| 4 |
Hb_002042_200 |
0.0965818234 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_000580_070 |
0.1081402436 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 6 |
Hb_002301_420 |
0.1088631053 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0011s05320g [Populus trichocarpa] |
| 7 |
Hb_003734_120 |
0.1105814247 |
- |
- |
PREDICTED: uncharacterized protein LOC105632812 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002182_050 |
0.1127615209 |
- |
- |
hypothetical protein CICLE_v10022654mg [Citrus clementina] |
| 9 |
Hb_118707_020 |
0.1167683086 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000380_210 |
0.1221936832 |
- |
- |
Cyclic nucleotide-gated channel 15 [Theobroma cacao] |
| 11 |
Hb_000276_030 |
0.1244617544 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 12 |
Hb_001623_400 |
0.1268047812 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_006637_040 |
0.127576742 |
- |
- |
phenylalanine ammonia-lyase 2 [Manihot esculenta] |
| 14 |
Hb_002749_070 |
0.1276386581 |
- |
- |
PREDICTED: uncharacterized protein LOC105635941 [Jatropha curcas] |
| 15 |
Hb_025254_020 |
0.129497893 |
- |
- |
PREDICTED: formate dehydrogenase, mitochondrial [Vitis vinifera] |
| 16 |
Hb_002597_010 |
0.1302611816 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Malus domestica] |
| 17 |
Hb_000487_240 |
0.1315662913 |
- |
- |
PREDICTED: CYSTM1 family protein B [Cucumis melo] |
| 18 |
Hb_001959_030 |
0.132846713 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004544_070 |
0.1347595186 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_188923_010 |
0.1357785283 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |