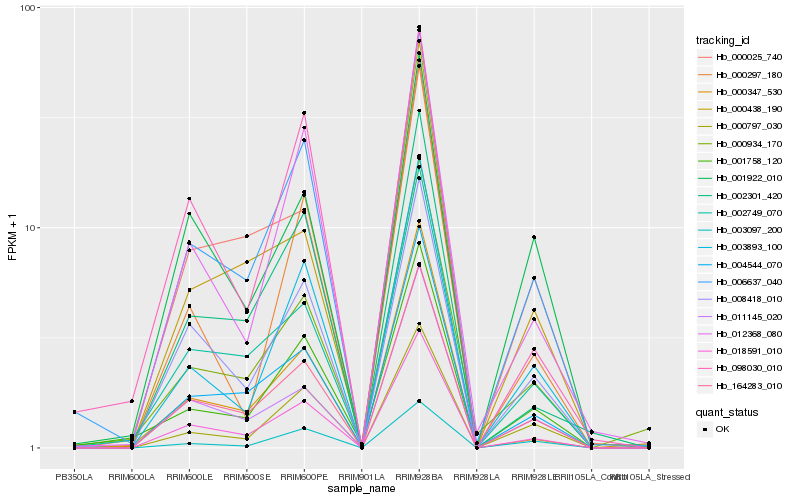
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_018591_010 |
0.0 |
- |
- |
kinase, putative [Ricinus communis] |
| 2 |
Hb_164283_010 |
0.0340029814 |
- |
- |
hypothetical protein RCOM_0725500 [Ricinus communis] |
| 3 |
Hb_008418_010 |
0.0648796824 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH93-like [Jatropha curcas] |
| 4 |
Hb_006637_040 |
0.0685644622 |
- |
- |
phenylalanine ammonia-lyase 2 [Manihot esculenta] |
| 5 |
Hb_004544_070 |
0.0741063425 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_002749_070 |
0.0818464276 |
- |
- |
PREDICTED: uncharacterized protein LOC105635941 [Jatropha curcas] |
| 7 |
Hb_011145_020 |
0.0858364663 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Pyrus x bretschneideri] |
| 8 |
Hb_002301_420 |
0.0885058786 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0011s05320g [Populus trichocarpa] |
| 9 |
Hb_012368_080 |
0.0919287809 |
- |
- |
PREDICTED: uncharacterized protein LOC105126277 [Populus euphratica] |
| 10 |
Hb_003893_100 |
0.0967923626 |
- |
- |
PREDICTED: uncharacterized protein LOC105648774 [Jatropha curcas] |
| 11 |
Hb_000347_530 |
0.109603885 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB114-like [Jatropha curcas] |
| 12 |
Hb_000797_030 |
0.1127643718 |
- |
- |
PREDICTED: leucoanthocyanidin dioxygenase [Jatropha curcas] |
| 13 |
Hb_001758_120 |
0.1133613586 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_098030_010 |
0.1168036741 |
- |
- |
hypothetical protein JCGZ_06251 [Jatropha curcas] |
| 15 |
Hb_000934_170 |
0.1196610682 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_003097_200 |
0.1235730805 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000297_180 |
0.1238472049 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |
| 18 |
Hb_000025_740 |
0.1244938682 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001922_010 |
0.1253404396 |
- |
- |
PREDICTED: ABC transporter G family member 8 [Jatropha curcas] |
| 20 |
Hb_000438_190 |
0.1300605012 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |