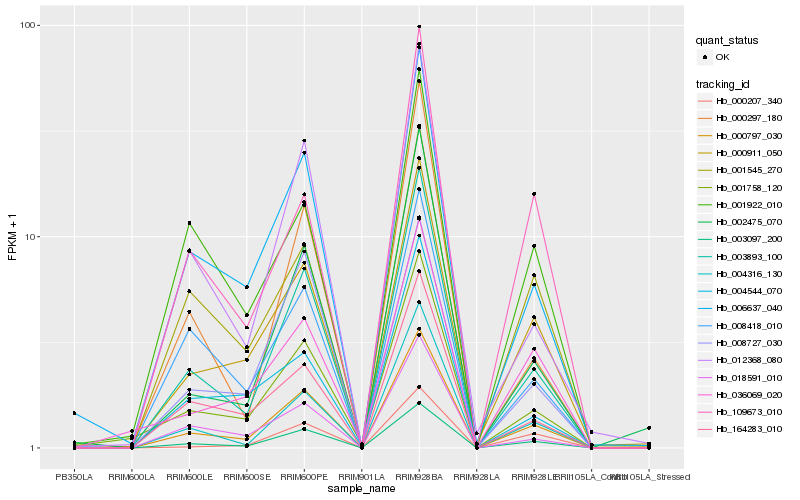
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000797_030 |
0.0 |
- |
- |
PREDICTED: leucoanthocyanidin dioxygenase [Jatropha curcas] |
| 2 |
Hb_003097_200 |
0.0211851062 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003893_100 |
0.0532211928 |
- |
- |
PREDICTED: uncharacterized protein LOC105648774 [Jatropha curcas] |
| 4 |
Hb_000911_050 |
0.0831135715 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 21 [Jatropha curcas] |
| 5 |
Hb_006637_040 |
0.0903548285 |
- |
- |
phenylalanine ammonia-lyase 2 [Manihot esculenta] |
| 6 |
Hb_004316_130 |
0.0952263167 |
- |
- |
PREDICTED: glutathione transferase GST 23-like, partial [Eucalyptus grandis] |
| 7 |
Hb_001758_120 |
0.1018306571 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_164283_010 |
0.1062340961 |
- |
- |
hypothetical protein RCOM_0725500 [Ricinus communis] |
| 9 |
Hb_012368_080 |
0.1112793084 |
- |
- |
PREDICTED: uncharacterized protein LOC105126277 [Populus euphratica] |
| 10 |
Hb_018591_010 |
0.1127643718 |
- |
- |
kinase, putative [Ricinus communis] |
| 11 |
Hb_001545_270 |
0.1148127608 |
- |
- |
laccase, putative [Ricinus communis] |
| 12 |
Hb_008418_010 |
0.1152723212 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH93-like [Jatropha curcas] |
| 13 |
Hb_000207_340 |
0.1197624122 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0013s11930g [Populus trichocarpa] |
| 14 |
Hb_002475_070 |
0.121233673 |
- |
- |
hypothetical protein POPTR_0017s02440g, partial [Populus trichocarpa] |
| 15 |
Hb_036069_020 |
0.1250633294 |
- |
- |
PREDICTED: receptor protein kinase-like protein At4g34220 [Jatropha curcas] |
| 16 |
Hb_000297_180 |
0.1315521638 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |
| 17 |
Hb_109673_010 |
0.1352577433 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 18 |
Hb_008727_030 |
0.1356077647 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004544_070 |
0.1377063632 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_001922_010 |
0.1379820457 |
- |
- |
PREDICTED: ABC transporter G family member 8 [Jatropha curcas] |