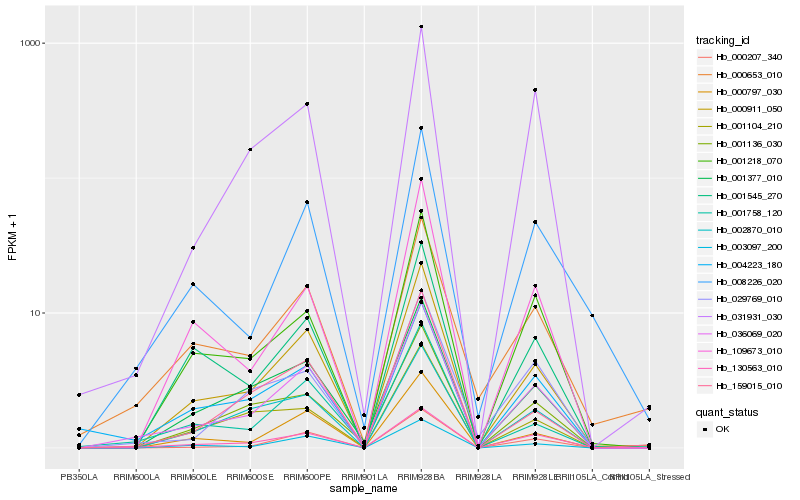
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_036069_020 |
0.0 |
- |
- |
PREDICTED: receptor protein kinase-like protein At4g34220 [Jatropha curcas] |
| 2 |
Hb_000911_050 |
0.0911396832 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 21 [Jatropha curcas] |
| 3 |
Hb_130563_010 |
0.0966624397 |
- |
- |
PREDICTED: ABC transporter C family member 3-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_001758_120 |
0.1203169284 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_001377_010 |
0.121069845 |
- |
- |
PREDICTED: transcription factor TRY-like [Jatropha curcas] |
| 6 |
Hb_000207_340 |
0.1238298518 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0013s11930g [Populus trichocarpa] |
| 7 |
Hb_000797_030 |
0.1250633294 |
- |
- |
PREDICTED: leucoanthocyanidin dioxygenase [Jatropha curcas] |
| 8 |
Hb_001218_070 |
0.1291732845 |
- |
- |
PREDICTED: sugar transport protein 5-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_001136_030 |
0.1311424609 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 25-like [Jatropha curcas] |
| 10 |
Hb_003097_200 |
0.1322285538 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004223_180 |
0.1350156515 |
- |
- |
PREDICTED: auxin-induced protein 15A [Jatropha curcas] |
| 12 |
Hb_031931_030 |
0.1372364722 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 13 |
Hb_008226_020 |
0.1422105675 |
- |
- |
hypothetical protein JCGZ_03321 [Jatropha curcas] |
| 14 |
Hb_001545_270 |
0.1442159659 |
- |
- |
laccase, putative [Ricinus communis] |
| 15 |
Hb_002870_010 |
0.1442498008 |
- |
- |
PREDICTED: uncharacterized protein LOC105649339 [Jatropha curcas] |
| 16 |
Hb_029769_010 |
0.1442610615 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 isoform X3 [Populus euphratica] |
| 17 |
Hb_109673_010 |
0.1475961257 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 18 |
Hb_159015_010 |
0.1476510427 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 19 |
Hb_000653_010 |
0.151359191 |
- |
- |
hypothetical protein CICLE_v10033238mg [Citrus clementina] |
| 20 |
Hb_001104_210 |
0.1552519619 |
- |
- |
hypothetical protein JCGZ_23620 [Jatropha curcas] |