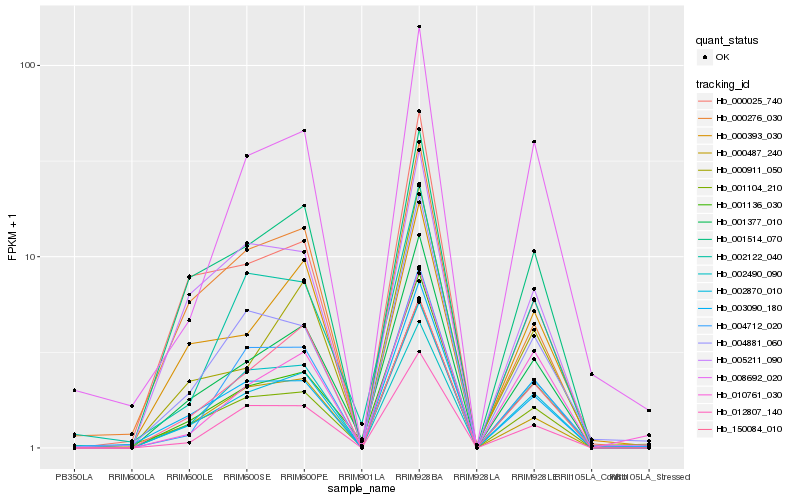
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002870_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649339 [Jatropha curcas] |
| 2 |
Hb_001377_010 |
0.0656064847 |
- |
- |
PREDICTED: transcription factor TRY-like [Jatropha curcas] |
| 3 |
Hb_001514_070 |
0.0820821337 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_001136_030 |
0.0881267338 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 25-like [Jatropha curcas] |
| 5 |
Hb_001104_210 |
0.0911467643 |
- |
- |
hypothetical protein JCGZ_23620 [Jatropha curcas] |
| 6 |
Hb_012807_140 |
0.0939125197 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0009s01270g [Populus trichocarpa] |
| 7 |
Hb_003090_180 |
0.1025098077 |
- |
- |
PREDICTED: uncharacterized protein LOC105646674 [Jatropha curcas] |
| 8 |
Hb_000487_240 |
0.1054479716 |
- |
- |
PREDICTED: CYSTM1 family protein B [Cucumis melo] |
| 9 |
Hb_004712_020 |
0.1059643408 |
- |
- |
hypothetical protein JCGZ_00782 [Jatropha curcas] |
| 10 |
Hb_005211_090 |
0.1104636825 |
- |
- |
PREDICTED: aluminum-activated malate transporter 4-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000393_030 |
0.1111341457 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 12 |
Hb_008692_020 |
0.1113246444 |
- |
- |
- |
| 13 |
Hb_004881_060 |
0.1167553068 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0005s22480g [Populus trichocarpa] |
| 14 |
Hb_000911_050 |
0.1221823259 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 21 [Jatropha curcas] |
| 15 |
Hb_002122_040 |
0.1233445006 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 16 |
Hb_000276_030 |
0.1260225434 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 17 |
Hb_010761_030 |
0.1267436115 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 18 |
Hb_002490_090 |
0.1300906906 |
- |
- |
PREDICTED: uncharacterized protein LOC105133003 [Populus euphratica] |
| 19 |
Hb_150084_010 |
0.1323121601 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Vitis vinifera] |
| 20 |
Hb_000025_740 |
0.1345429297 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12 isoform X1 [Jatropha curcas] |