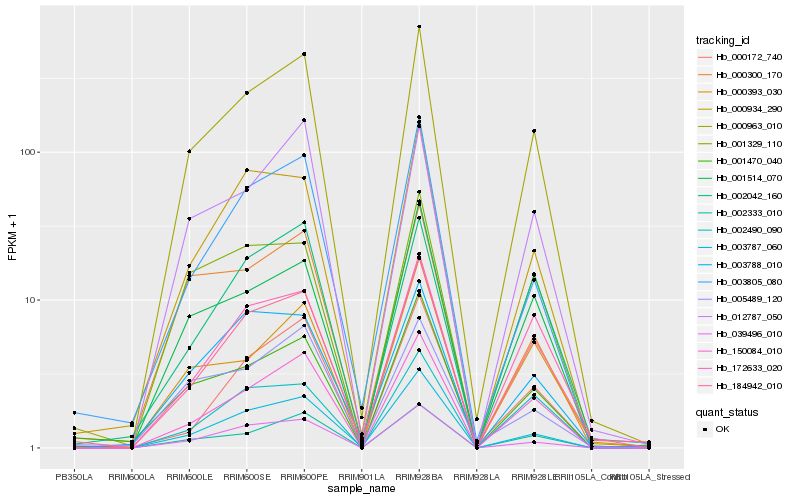
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000963_010 |
0.0 |
- |
- |
PREDICTED: aquaporin PIP2-4 [Jatropha curcas] |
| 2 |
Hb_150084_010 |
0.062117603 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Vitis vinifera] |
| 3 |
Hb_002333_010 |
0.065893381 |
- |
- |
hypothetical protein JCGZ_18834 [Jatropha curcas] |
| 4 |
Hb_003787_060 |
0.0856984903 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 5 |
Hb_184942_010 |
0.0867961209 |
- |
- |
STS14 protein precursor, putative [Ricinus communis] |
| 6 |
Hb_039496_010 |
0.0880636542 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 7 |
Hb_012787_050 |
0.1035911589 |
- |
- |
PREDICTED: glutathione S-transferase U18-like [Jatropha curcas] |
| 8 |
Hb_001514_070 |
0.1075482795 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_172633_020 |
0.1135473335 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g56140 [Populus euphratica] |
| 10 |
Hb_003805_080 |
0.113634605 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 11 |
Hb_002042_160 |
0.1147871135 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 12 |
Hb_003788_010 |
0.1157023531 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 13 |
Hb_000934_290 |
0.115873339 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001470_040 |
0.1178200837 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 3 [Jatropha curcas] |
| 15 |
Hb_000172_740 |
0.1212671427 |
- |
- |
HXXXD-type acyl-transferase family protein, putative [Theobroma cacao] |
| 16 |
Hb_000300_170 |
0.1223208996 |
- |
- |
PREDICTED: uncharacterized protein LOC105632565 [Jatropha curcas] |
| 17 |
Hb_002490_090 |
0.1224274321 |
- |
- |
PREDICTED: uncharacterized protein LOC105133003 [Populus euphratica] |
| 18 |
Hb_001329_110 |
0.1225653864 |
- |
- |
PREDICTED: uncharacterized protein LOC105642675 [Jatropha curcas] |
| 19 |
Hb_000393_030 |
0.1227082574 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 20 |
Hb_005489_120 |
0.1231529809 |
- |
- |
leucine rich repeat receptor kinase, putative [Ricinus communis] |