| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
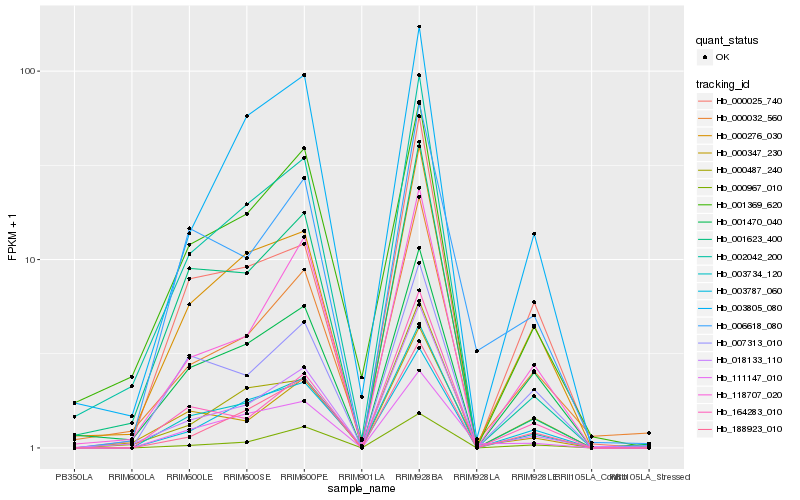
Hb_003734_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632812 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000276_030 |
0.0827190977 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 3 |
Hb_003787_060 |
0.1018111835 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 4 |
Hb_188923_010 |
0.1104160484 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 5 |
Hb_018133_110 |
0.1105814247 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 6 |
Hb_007313_010 |
0.1107467767 |
- |
- |
hypothetical protein JCGZ_23398 [Jatropha curcas] |
| 7 |
Hb_002042_200 |
0.1118906542 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_001623_400 |
0.1153429498 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_118707_020 |
0.1177093779 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003805_080 |
0.1183365752 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 11 |
Hb_001369_620 |
0.1204438297 |
- |
- |
PREDICTED: protein EXORDIUM-like 3 [Jatropha curcas] |
| 12 |
Hb_000487_240 |
0.1210033644 |
- |
- |
PREDICTED: CYSTM1 family protein B [Cucumis melo] |
| 13 |
Hb_111147_010 |
0.1232300229 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000032_560 |
0.1264095532 |
- |
- |
hypothetical protein JCGZ_15037 [Jatropha curcas] |
| 15 |
Hb_000025_740 |
0.1271029271 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001470_040 |
0.1297185756 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 3 [Jatropha curcas] |
| 17 |
Hb_000347_230 |
0.130141369 |
- |
- |
inter-alpha-trypsin inhibitor heavy chain, putative [Ricinus communis] |
| 18 |
Hb_000967_010 |
0.1312025351 |
- |
- |
hypothetical protein JCGZ_08336 [Jatropha curcas] |
| 19 |
Hb_164283_010 |
0.1312957729 |
- |
- |
hypothetical protein RCOM_0725500 [Ricinus communis] |
| 20 |
Hb_006618_080 |
0.1313519363 |
- |
- |
PREDICTED: non-specific lipid-transfer protein [Jatropha curcas] |